Evolving therapies have allowed the use of sperm from men with spermatogenic compromise, obstructive azoospermia, and sperm functional deficiency, enabling these men to procreate when unable to do so naturally. The genetic basis of only a portion of these conditions is known and research must be pursued into the genetic underpinnings of those that have not yet been delineated. Education and provision of information to patients is the responsibility of all involved in the care of men with reproductive failure. The author concentrates on some of the known causes of nonobstructive azoospermia and obstructive azoospermia with a well-established genetic cause such as congenital bilateral absence of the vas deferens.
Darwinian evolution is most concerned with adaptation and change of morphology or phenotype (ie, survival of the fittest). The lineage of almost all current species can be traced back to ancestral forms by similarity in form and function and also by molecular signatures. We can outline our human roots back millions of years by these methods and know that we have attained our present form through some incredibly remarkable changes, most notably brain development. Evolution and adaptational modification continue unabated as the environment changes; stressors are altered and new niches open up, whereas others close down. But all along, nature and evolution have been, and still are, paying close attention to another function: reproduction . Transformation of the genome, at the genetic or epigenetic level, is what underlies the process of evolution. This transformation may take place in a slow and steady fashion or in quantum leaps, relatively speaking. If an advantageous variation in the genome occurs that, for example, allows the proband organism to survive better in a particular environment but completely impairs reproductive capability, that variation will not be transmitted to the next generation and that beneficial genetic “mishap” will be lost. If that same advantageous variation did not alter reproductive capacity, it will indeed be passed along to benefiting recipient offspring who will also have an improved survival, allowing them to procreate more successfully and affecting the future offspring of that lineage. If nature is all about the transmitting of DNA, then survival and reproduction are equally important and, therefore, it would be distinctly unusual for any surviving organism from any species to have an impaired reproductive ability. Survival and evolution depend on procreation. One should look at human reproductive failure the same way; unless we can prove otherwise, we should assume impaired fertility has a genetic basis and we should inform couples about this. Our therapies to treat severe male factor infertility have far outstripped our knowledge of the reasons behind that failure. Intracytoplasmic sperm injection (ICSI) is a process whereby a single sperm is directly injected into a single oocyte, allowing the use of sperm from the testis, even in the most impaired situations in which only a handful of sperm are produced secondary to near-total spermatogenic failure. Is the use of this sperm safe? What are the short- and long-term consequences? What will be the reproductive capability of the offspring? It is a large human experiment, the subjects of which are not the couples we are helping, but the children we are creating. We are bypassing the natural selection process that forms the basis of evolutionary selection.
In addition, the male reproductive axis of every species is designed to be quantitatively and qualitatively maximized; reserve sperm production potential is not needed. Finally, meiosis occurs in only two places, oogenesis and spermatogenesis, and for each, a cadre of genes has evolved just for this purpose; these genes may be dysfunctional with no somatic effects, just defective egg or sperm production. These last two points fully describe many men with severe spermatogenic inadequacy; they are phenotypically healthy in all respects, except for reduced or absent sperm production . In this article, the author concentrates on some of the known causes of nonobstructive azoospermia (NOA) (spermatogenic failure) and obstructive azoospermia (OA) with a well-established genetic cause such as congenital bilateral absence of the vas deferens (CBAVD). These, in combination, represent most of what will be seen in an infertility practice. Finally, mention will be made of the syndromes that may be rarely encountered that affect sperm form (globozoospermia and fibrous sheath dysplasia) and sperm function (immotile cilia syndrome).
Nonobstructive azoospermia: Y chromosomal microdeletions
The basic molecular geography of the Y chromosome is important to appreciate before delving into the clinical consequences of Y chromosomal microdeletions. The Y chromosome is approximately 60 million base pairs in length, equally divided between the euchromatic and heterochromatic regions . At the ends of both the short (Yp) and long (Yq) arms are “pseudoautosomal regions” that pair and recombine with like regions on the X chromosome . One of the most important genes in the cascade that determines the fate of the bipotential gonad, sex-determining region Y (SRY), is located on Yp . Between the two pseudoautosomal regions at the ends of the Y is unique chromosomal material not represented elsewhere in the genome, which is termed the “male-specific Y” (MSY), composing approximately 95% of the Y chromosome ( Fig. 1 ) . As such, it is nonrecombining and was previously known as nonrecombining Y (NRY). Within MSY, multiple genes are sprinkled throughout, most involved with spermatogenesis yet still poorly characterized at this point . Examples of these genes include USP9Y, DBY, DAZ, RBMY1 , and BPY2 . Within the euchromatic portion of the long arm of Y are interspersed eight palindromic sequences, labeled in order, P8 to P1, beginning closest to the centromere. Each palindrome is of a different length but all have a central short base pair core from which mirror-image, identical stretches spread outwards, reading exactly the same but, of course, in opposite directions. Certain palindromes are made up of subsegments or amplicons, of which at least two copies exist (one on each arm) but occasionally of which more than two copies are present (spatially separate in a different palindrome and reading in either the same orientation [direct] or in the opposite direction [inverted]) . It is thought that this repetitive-style molecular organization of the MSY has evolved as a mechanism for maintaining the presence and fidelity of the MSY because the genome does not have a counterpart where correction and renewal take place during mitotic pairing and meiotic recombination. However, this same molecular arrangement may also rarely lead to ectopic homologous recombination during Y chromosomal replication, a process whereby two spatially distanced subsegments “stick” together, with resultant loss of all chromosomal material in the intervening portion . Such losses are referred to as microdeletions because they are not visible by cytogenetic analysis. If these microdeletions occur, then the resident gene population is also lost. In summary, most of the Y chromosome is unique, the organization is unusual, genes involved in the spermatogenic process reside in MSY, long segments of the Y may be lost if ectopic homologous recombination occurs, and, if this recombination occurs, any genes that live in these regions will also disappear.
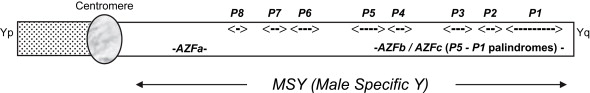
The description of the molecular anatomy of the Y is important to understand vis-à-vis clinical male reproduction because several genes sprinkled along the length of proximal Yq are either necessary or helpful for optimal spermatogenesis. It was recognized in 1976 that cytogenetically recognizable gross deletions and anomalies of the Y chromosome could be found in men with spermatogenic failure. However, as the molecular anatomy of the Y chromosome was being elucidated in the mid-1990s and beyond, specific microdeletions of the Y chromosome were being discovered in men with sperm production deficiency. The original acronym nomenclature arose at this time when it was thought that only three of these spatially and topographically distinct microdeleted regions existed, AZF (azoospermia factor) a , AZFb , and AZFc . A Y chromosomal microdeletion assay is a blood test, readily available, that can determine whether one of the clinically important microdeletions is present and it should be obtained in all NOA or severely oligospermic men, before either using ejaculated sperm in conjunction with ICSI or surgically attempting to harvest testis sperm.
The AZFa region is located in proximal Yq, is 792 kilobases (kb) in length, and contains DDX3Y (also known as DBY ) and USP9Y , two genes felt to be important in the spermatogenic process . For example, DDX3Y is 16.3 kb in length, generates an ATP-dependent RNA helicase that shuttles between the nucleus and cytoplasm, and may play a part in the later stages of spermatogenesis . USP9Y may play less of a quantitative role in spermatogenesis than DDX3Y ; mutations in USP9Y have been described in men with some spermatogenic potential . However, flanking the genomic material where DDX3Y and USP9Y call home are two 10-kb endogenous retroviral elements, HERV15yq1 and HERV15yq2 . Ectopic homologous recombination may occur between these two sequences, with resultant loss of the intervening material, including DDX3Y and USP9Y . An AZFa microdeletion is found to be the proximate cause in approximately 1% of NOA men. Spermatogenic failure is the ultimate clinical consequence and the literature suggests that sperm will not be found in the tissue on testis sperm extraction (TESE). Therefore, detection of an AZFa microdeletion dictates that TESE not be performed because the prognosis for sperm retrieval is grave .
The AZFb and AZFc regions are located further along Yq in the P5 through P1 genomic span. In actuality, multiple possible sites of microdeletion may arise in this stretch. Known by various names, however, such as AZFb and AZFc , they are simply different microdeletions of different lengths, occurring at different frequencies, and with different proximal and distal end points, but all within the boundaries mentioned earlier and all a consequence of ectopic homologous recombination. AZFb and AZFc appeared to be distinct and nonoverlapping on Yq in the early days of sequencing of the Y chromosome. Precise definition of the P5 to P1 interval has shown, however, that the AZFb and AZFc regions are indeed overlapping and simply represent different sites of ectopic homologous recombination within this expanse .
AZFc is the most common microdeleted region found, happening de novo in approximately 1:4000 men overall, and found in 13% of azoospermic and 6% of severely oligospermic men . The AZFc region spans 3.5 megabases (Mb), beginning in the distal aspect of the P3 palindrome and extending into the P1 palindrome . The AZFc region microdeletion is also known as b2/b4 because two of the four blue amplicons (as colorized in the Kuroda-Kawaguchi article, 229 kb in length) flank this 3.5-Mb expanse and are the sequences that undergo ectopic homologous recombination; the resulting loss of intervening genomic material is what we define as an AZFc microdeletion. An AZFc microdeletion event removes several genes that inhabit this region, perhaps most importantly, the four copies of DAZ that exist here . DAZ encodes an RNA-binding protein expressed primarily in spermatogonia, possibly activating silent mRNAs during meiosis . The genes within the AZFc region are not critical for meiotic recombination but “in the absence of the AZF region, the transient zygotene stage is extended, and chromosome condensation is reduced” .
Oates and colleagues have provided the most relevant clinical correlations to AZFc microdeletions, as follows. An AZFc microdeletion quantitatively impairs spermatogenesis. Infertility and sterility are the rule, although natural paternity has been reported . It is extremely rare to find sperm density greater than 5 ×106/mL. In their sample of 42 men, 38% were severely oligospermic and 62% were azoospermic. Of the azoospermic men, 67% had some level of spermatogenesis noted on testis biopsy or TESE. Critical to remember, however, is that 19% of the overall group did not have sperm available from either the ejaculate or testis tissue. AZFc -microdeleted men do not appear to have any other health or testis-specific consequences; the genes lost appear to be important only in spermatogenesis. Nearly all AZFc microdeletions are de novo (the father is not microdeleted himself, but the Y chromosome in the sperm that he produced and that fertilized the egg that became the patient had suffered a microdeletion event). Spermatogenic potential, at whatever low level it might be, appears to be stable over time. No historical or physical findings, or hormonal predictive factors, can forecast whether sperm will be found in the ejaculate, in the testis tissue only, or not at all. When spermatozoa from an AZFc -microdeleted man are used in conjunction with intracytoplasmic sperm injection, they appear to work well; quality is preserved . The children of AZFc -microdeleted men are somatically healthy but all male offspring will harbor an AZFc microdeletion and the spectrum of reproductive impairments is similar to that found in de novo cases, with their level of ultimate spermatogenesis not necessarily the same as their fathers’ ( Fig. 2 ) . If a Y chromosomal microdeletion assay defines an AZFc microdeletion, the couple may choose not to use his sperm, may choose to use it for ICSI, or may choose to use preimplantation genetic screening such that only female embryos will be transferred, thereby eliminating the propagation of an AZFc microdeletion and its consequent infertility/sterility .









