Humans are host to complex microbial communities previously termed normal flora and largely overlooked. However, resident microbes contribute to both health and disease. Investigators are beginning to define microbes that contribute to the development of gastrointestinal malignancies and the mechanisms by which this occurs. Resident microbes can induce inflammation, leading to cell proliferation and altered stem cell dynamics, which can lead to alterations in DNA integrity and immune regulation and promote carcinogenesis. Studies in human patients and rodent models of cancer have identified alterations in the microbiota of the stomach, esophagus, and colon that increase the risk for malignancy.
Key points
- •
The risk of developing gastric cancer is multifactorial, and the microbiota has been identified as an important contributing factor.
- •
Colon cancer risk is modified by the gastrointestinal tract microbiota and environmental exposures, including diet, in addition to known genetic factors.
- •
With no single microbial causative agent identified, it is likely that an overall disturbance in the composition/metabolism of the colonic microbiota can promote cancer development.
Introduction
In the last 2 decades, there has been a remarkable shift in identifying and understanding the multitude of microbes that colonize the human body. Previously, the normal flora was thought to be largely a silent passenger, only declaring itself when it traveled outside of its usual niche. However, it is now recognized that the microbiome, which is composed of bacteria, archaea, eukaryotes, and viruses, plays a key role in health and disease. Bacteria are the most abundant and well studied. The gastrointestinal (GI) microbiome is molded from birth by a multitude of interactions that can be distinct, such as the host genetic background, or variable, including diet, antibiotics, and other environmental exposures.
Cancer is the second leading cause of death in the United States, and GI cancers represent a leading cause of morbidity and mortality. Although genetic factors leading to an increased risk of cancer have been identified, such as adenomatous polyposis coli ( APC ) mutations that lead to familial adenomatous polyposis and E-cadherin ( CDH1 ) mutations that lead to hereditary diffuse-type gastric cancer (GC), these mutations do not account for most cases. In addition, the association of microbial infections with the risk for cancer development is well documented, including Helicobacter pylori with GC and hepatitis viruses with liver cancer. Even nonpathogenic GI tract microbes, once considered inert, have been found to play a role in chronic inflammation, altering cell proliferation and stem cell dynamics, and altering immune surveillance mechanisms. The focus of this review is the role of the GI microbiome in the development of gastric and colonic malignancies with a brief discussion of esophageal malignancy.
Introduction
In the last 2 decades, there has been a remarkable shift in identifying and understanding the multitude of microbes that colonize the human body. Previously, the normal flora was thought to be largely a silent passenger, only declaring itself when it traveled outside of its usual niche. However, it is now recognized that the microbiome, which is composed of bacteria, archaea, eukaryotes, and viruses, plays a key role in health and disease. Bacteria are the most abundant and well studied. The gastrointestinal (GI) microbiome is molded from birth by a multitude of interactions that can be distinct, such as the host genetic background, or variable, including diet, antibiotics, and other environmental exposures.
Cancer is the second leading cause of death in the United States, and GI cancers represent a leading cause of morbidity and mortality. Although genetic factors leading to an increased risk of cancer have been identified, such as adenomatous polyposis coli ( APC ) mutations that lead to familial adenomatous polyposis and E-cadherin ( CDH1 ) mutations that lead to hereditary diffuse-type gastric cancer (GC), these mutations do not account for most cases. In addition, the association of microbial infections with the risk for cancer development is well documented, including Helicobacter pylori with GC and hepatitis viruses with liver cancer. Even nonpathogenic GI tract microbes, once considered inert, have been found to play a role in chronic inflammation, altering cell proliferation and stem cell dynamics, and altering immune surveillance mechanisms. The focus of this review is the role of the GI microbiome in the development of gastric and colonic malignancies with a brief discussion of esophageal malignancy.
Gastric cancer
Gastric adenocarcinoma is the third leading cause of cancer-related death in the world. In developed countries, the incidence of gastric adenocarcinoma has significantly decreased over the past century ; however, the incidence rates of both proximal gastric and gastroesophageal junction adenocarcinomas have increased in both the United States and Europe. Chronic infection with H pylori is the strongest known risk factor for developing gastric adenocarcinoma.
Helicobacter pylori
H pylori is a gram-negative bacteria that selectively colonizes the gastric epithelium. Infection is usually acquired in childhood and, in the absence of combination antibiotic therapy, can persist for the lifetime of the host. H pylori has colonized humans for almost 100,000 years ; approximately half of the world’s population is infected with H pylori , promoting speculation that H pylori is an endogenous member of the gastric microbiota. Between 1% and 3% of H pylori –colonized persons develop gastric adenocarcinoma ; factors that play a role in the pathologic outcome of H pylori infection are varied, including strain-specific bacterial constituents; host genetic factors; environmental influences, including diet; and alterations in the host microbiota.
Bacterial and host factors affecting the propensity toward gastric cancer
One H pylori virulence factor that influences GC risk is the cag pathogenicity island, which contains genes encoding proteins that form a type IV bacterial secretion system. Another H pylori virulence factor linked to the development of GC is the secreted vacuolating cytotoxin A (VacA). All H pylori strains contain vacA , but there are considerable differences in vacA sequences among strains. Strains containing type s1, i1, or m1 alleles within the 5′ region of the gene are highly associated with GC. Host polymorphisms in interleukin ( IL ) -1β and tumor necrosis factor ( TNF ) -α as well as environmental factors, such as a high-salt diet and low iron levels, in the context of H pylori infection also influence gastric carcinogenesis.
Although H pylori infection is the strongest identified risk factor for developing GC, clinical trials suggest that other gastric microbiota constituents may influence disease progression. Antibiotic therapy directed against H pylori was reported to significantly decrease the incidence of GC in a 15-year follow-up study of 3365 subjects. Of note, more than 50% of the antibiotic-treated individuals remained colonized by H pylori at the 15-year follow-up. These findings suggest that antibiotic treatment may attenuate the development of GC by inducing alterations in the non– H pylori microbiota.
The stomach microbiota in gastric pathogenesis
The stomach harbors a large and diverse bacterial community ranging from 10 1 to 10 3 colony-forming units per gram, which may influence gastric homeostasis and disease in conjunction with H pylori infection.
The composition of the gastric microbiome in H pylori –negative individuals is highly diverse ( Fig. 1 ). Sequencing of DNA isolated from human gastric biopsies identified 128 phylotypes within 8 bacterial phyla of which Proteobacteria, Firmicutes, Bacteroidetes, Fusobacteria, and Actinobacteria were the most abundant. Using a newer technology, tagged 454 pyrosequencing, analysis of H pylori –negative biopsy samples identified 262 phylotypes representing 13 phyla. These findings lend further support to the gastric microbiota being highly diverse, despite significant variability in the microbial composition between individuals. In contrast, the microbiota among H pylori –infected individuals is much more uniform; H pylori represents the most abundant phylotype present in the stomach of H pylori –positive persons. H pylori DNA accounted for 93% to 97% of all sequence reads in H pylori –positive persons and a total of 33 phylotypes were detected, more than 200 less than in H pylori –negative persons. Taken together, these data suggest that H pylori colonization dramatically alters gastric microbiota diversity (see Fig. 1 ). Characterization of the human gastric microbiota using DNA microarrays detected 44 phyla with 4 dominant phyla: Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes. Using this method, infection with H pylori was shown to increase the relative abundance of non– H pylori Proteobacteria, Spirochaetes, and Acidobacteria and decrease the relative abundance of Actinobacteria, Bacteroidetes, and Firmicutes compared with uninfected stomachs. H pylori infection accounted for 28% of the variance in the microbiota; however, the bacterial communities in both H pylori –negative and –positive individuals remained highly complex.
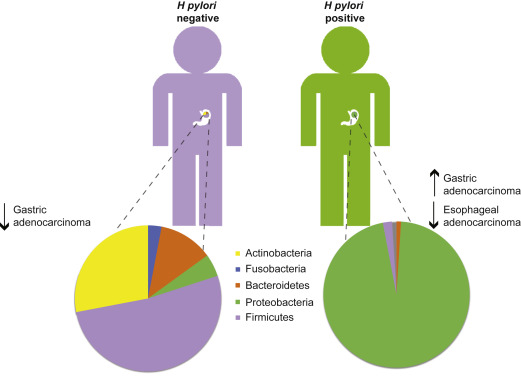
Studies examining differences in microbial composition and outcomes of GC are more limited. Development of atrophic gastritis, which induces hypochlorhydria due to parietal cell loss, is a key step in the histologic progression to intestinal-type GC and can lead to overgrowth of non- Helicobacter microbiota, which may promote the progression towards GC. Two recent studies have independently identified that proton pump inhibitor use may detrimentally alter the gut microbiota.
When comparing the microbiota of 10 patients with GC to 5 dysplastic controls, the microbiota of patients with GC was found to be equally as diverse as dysplastic patients. Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria were identified. The microbiota was predominately composed of species of Streptococcus , Lactobacillus , Veillonella , and Prevotella . H pylori were present in relatively low abundance. More recently, pyrosequencing has been used to compare the gastric microbiota in persons with chronic gastritis, intestinal metaplasia, and GC. Pyrosequencing identified 10 bacterial phyla, and significant differences were observed in both the composition and diversity of the gastric microbiota in the histologic progression towards GC. Bacilli and members of the Streptococcaceae family were significantly enriched in GC samples compared with chronic gastritis and intestinal metaplasia samples, whereas Epsilonproteobacteria and Helicobacteraceae family members were decreased.
An interesting new study compared the gastric microbiota of subjects from 2 Colombian populations: one at high-risk, Tuquerres, and one at low-risk, Tumaco, of developing GC. Despite high variability in the microbial composition between individuals, significant correlations were found with the town of origin. Two operational taxonomic units, Leptotrichia wadei , which is associated with necrotizing enterocolitis and bacteremia, and a Veillonella sp , were significantly more abundant in Tuquerres. In the low-risk region of Tumaco, 16 operational taxonomic units, including a Staphylococcus sp , which is considered a constituent of the normal human microbiota, were significantly more abundant. These results provide a tantalizing opportunity to manipulate the microbiota of animal models to more closely represent the microbiota of either the high-risk or low-risk populations of Colombia and determine key players in cancer development.
Animal models to study the microbiome and gastric cancer
Inbred mice with defined genotypes are commonly used to model carcinogenesis; however, this can be limited by uncontrolled microbial diversity within mice despite identical genetic backgrounds. To counter this, gnotobiotic mice allow for controlling the microbiome and adding back individual or collections of microorganisms.
Similar to humans, the most abundant phylotypes in the mouse stomach are Bacteroidetes, Firmicutes, Proteobacteria, and Actinobacteria ; infection of mice with H pylori can alter the gastric microbiota. H pylori infection induces gastritis in mice, and following H pylori infection for 2 months the gastric microbiota in specific pathogen free (SPF) mice harbored reduced numbers of Lactobacillus species and increased bacterial diversity. An independent study, however, found that both acute and chronic infection of SPF C57BL/6 mice with H pylori failed to cause significant shifts in the gastric microbial composition. It is possible that the inherent gastric microbial diversity of SPF mice may play a role in the outcome of H pylori infection.
INS-GAS mice are genetically predisposed to GC, and chronic interaction between H pylori and the gastric microbiota was found to influence disease progression in this model. In SPF INS-GAS mice, GC spontaneously developed. However, in germfree (GF) INS-GAS mice, cancer was slower to develop. Moreover, H pylori -infected GF INS-GAS mice developed less severe lesions and were slower to progress to GI intraepithelial neoplasia than H pylori– infected SPF INS-GAS mice. A detailed analysis using 454 sequencing of partial 16S ribosomal DNA amplicons revealed specific differences in phyla between H pylori –infected and uninfected SPF INS-GAS mice. H pylori colonization led to an expansion in the proportion of Firmicutes and decreased numbers of Bacteroidetes while causing an overall increase in species diversity. In fact, only 3 species of commensal bacteria (ASF356 Clostridium species, ASF361 Lactobacillus murinus , and ASF519 Bacteroides species ) were required to promote gastric neoplasia in H pylori –infected GF INS-GAS mice to the same extent as that reported in H pylori –infected SPF INS-GAS mice.
Esophageal adenocarcinoma and the microbiome
The incidence of esophageal adenocarcinoma has been increasing rapidly in developed countries over the past 40 years; this coincides with a decreasing incidence of H pylori infection and GC, suggesting that gastric colonization with H pylori may be protective against esophageal adenocarcinoma. This protection could reflect inhibition of acid secretion via enhanced production of IL-1β and TNF-α in response to H pylori or through loss of parietal cells in atrophic gastritis. Alternatively, changes in the gastric microbiota resulting from the loss of H pylori may increase the risk for an individual to develop esophageal cancer (see Fig. 1 ).
The esophageal microbiome is altered during inflammation and metaplasia. Using a 16S rRNA gene survey, 2 types of microbiota, termed type I and type II, were identified in the esophagus. The type I microbiome was dominated by gram-positive bacteria and the genus Streptococcus , whereas the type II microbiome was composed of a higher percentage of gram-negative bacteria, with the phyla Bacteroidetes, Proteobacteria, Fusobacteria, and Spirochaetes being the most abundant. The type II microbiome correlated with the histologic presence of esophagitis and Barrett’s esophagus, whereas the type I microbiome was associated with a histologically normal esophagus.
In a recent study, 30 esophageal adenocarcinoma cases were compared with 39 control subjects using cultured biofilms. In control subjects, 56 species belonging to 19 genera were detected, whereas in esophageal adenocarcinoma, 73 species from 23 genera were identified. Despite finding more species in esophageal adenocarcinoma than controls, no statistical differences were reported. These findings provide an important framework for more detailed future studies delineating the structure and function of the esophageal microbiome and disease.
Colorectal cancer
Colorectal cancer (CRC) is the third leading cause of cancer mortality in the United States, and the risk of CRC increases with age. Most of the cases are sporadic; however, up to 25% of patients have a family history of CRC but no evidence of an identified inherited syndrome. This finding underscores the complex interaction of multiple genetic and epigenetic events contributing to CRC pathogenesis. The initiation of CRC can be due to mutations in tumor-suppressor genes, such as APC , catenin ( cadherin-associated protein ) beta 1 , tumor protein p53 , and the oncogene Kirsten rat sarcoma viral oncogene homolog , leading to a growth advantage in colonic epithelial cells progressing to adenomas and cancer. Although these genetic mutations have clearly been linked to CRC development, the steps leading to the accumulation of these mutations and other epigenetic changes are not fully known. In addition to genetic alterations, microbial and environmental factors, including diet and lifestyle, have been shown in recent studies to play a role in promoting CRC.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree






