Emerging molecular tools promise to extend the diagnostic reach of the endoscopist and open doors to population screening for gastrointestinal (GI) cancers. This review briefly addresses biological considerations in marker detection and types of markers, highlights examples of tools under development at each organ site, and appraises the possibility of universal GI cancer screening. The outlook is positive, but further technical refinement and rigorous clinical validation are needed before most of these new approaches are ready for clinical application.
Key points
- •
Gastrointestinal (GI) malignancies account for 40% of all cancer deaths globally, but most types remain unscreened.
- •
With recent technology advances, new molecular screening tools under development could open doors for early detection of all GI cancers.
- •
Adjunctive molecular tests also have potential to extend the reach of the endoscopist for greater accuracy in detecting GI neoplasms.
Introduction
Globally, gastrointestinal (GI) malignancies account for roughly 40% of all cancer deaths. In the United States, upper GI cancers kill twice as many as does colorectal cancer (CRC); but, unlike CRC, rates for cancers of the pancreas, liver, and esophagus are increasing. This year, pancreatic cancer surpasses breast as the third most common cancer killer and by 2020 passes CRC as the second most common. By 2030, hepatoma may overtake CRC as the third deadliest cancer. Currently, most upper GI cancers present symptomatically at a late stage and are among the most lethal cancers. These alarming trends are calls for action.
Effective early detection methods are needed desperately. However, with a few exceptions, screening for upper GI cancers in most countries has not been pursued because of the relatively low prevalence of cancers at individual sites and lack of cost-effective screening tools.
This overview briefly examines molecular approaches as potential screening solutions to the challenge of GI cancer. It summarizes the types of molecular markers being considered, gives examples of molecular tools under development, and appraises the future role of molecular methods for universal detection of GI cancers. Although there are obvious implications on downstream endoscopic evaluation and management, these important clinical aspects are not addressed.
Introduction
Globally, gastrointestinal (GI) malignancies account for roughly 40% of all cancer deaths. In the United States, upper GI cancers kill twice as many as does colorectal cancer (CRC); but, unlike CRC, rates for cancers of the pancreas, liver, and esophagus are increasing. This year, pancreatic cancer surpasses breast as the third most common cancer killer and by 2020 passes CRC as the second most common. By 2030, hepatoma may overtake CRC as the third deadliest cancer. Currently, most upper GI cancers present symptomatically at a late stage and are among the most lethal cancers. These alarming trends are calls for action.
Effective early detection methods are needed desperately. However, with a few exceptions, screening for upper GI cancers in most countries has not been pursued because of the relatively low prevalence of cancers at individual sites and lack of cost-effective screening tools.
This overview briefly examines molecular approaches as potential screening solutions to the challenge of GI cancer. It summarizes the types of molecular markers being considered, gives examples of molecular tools under development, and appraises the future role of molecular methods for universal detection of GI cancers. Although there are obvious implications on downstream endoscopic evaluation and management, these important clinical aspects are not addressed.
Mechanisms, media, and markers
The concept of molecular screening is based on measurement of tumor-derived markers in readily accessible media. The promise of this approach lies in its ease for patients and potential to open doors for cost-effective screening of multiple GI cancers with a single test. Several key biological and technical elements must come together for molecular screening to be feasible. First, markers must reliably enter and remain in the target medium to be detectable at earliest cancer stage and, ideally, with precancer. Second, assays with sufficient technical sensitivity are needed to detect tumor markers in low abundance. And, third, highly discriminant markers must be identified that are positive when a GI neoplasm is present but otherwise remain negative, and such markers would ideally also predict the anatomic site of the primary GI tumor. Fueled by rapid technical advances, there is intense academic interest in this approach and increasing development by industry.
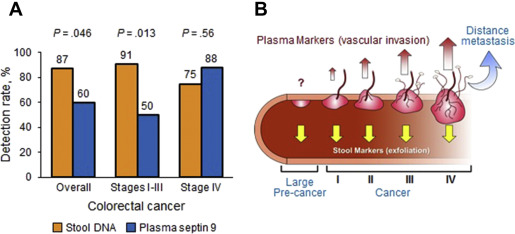
With GI cancers there exists the unique opportunity for noninvasive screening by marker detection either in blood or, because of their shared property of luminal exfoliation, in stool. A central obstacle with blood testing has been the difficulty detecting earliest stage cancer and precancer. Tumor markers are detected readily from circulation with later stage cancer, but are often below detection limits with stage I cancers and precancers. In contrast, CRC and precancerous polyps exfoliate molecular markers abundantly into stool. A direct comparison between stool and plasma testing of DNA markers in paired samples showed substantially higher marker levels and diagnostic yield with stool than with plasma ( Fig. 1 A ), which prompted a conceptual model suggesting that the biological mechanism of marker release by exfoliation favors early stage detection over that by vascular invasion ( Fig. 1 B). It may be possible to overcome the biological constraints of limited vascular invasion in early stage disease by exploiting potential alternative mechanisms of marker entry into blood, such as by release of exosomes (nanovesicles emitted from surface of tumor cells and containing proteins, RNA, and DNA) or phagocytosis by circulating macrophages, or simply by improved analytical sensitivity to detect the associated low plasma levels of tumor markers. With stool testing, it is unclear how effectively markers exfoliated from upper GI tumors can be recovered after traversing the digestive gauntlet. Model systems suggest that the small quantities of cells estimated to be shed from upper GI tumors can be detected in stool using sensitive techniques, and pilot case-control studies have demonstrated that it is possible to detect tumor markers in stool from patients with known upper GI cancer.
A comprehensive summary of specific candidate tumor markers is beyond the scope of this clinical review. The major classes of markers include intact tumor cells and cellular constituents including proteins, RNA, and DNA; each class has diagnostic advantages and disadvantages.
Whole Tumor Cells
Intact tumor cells may be recovered from the circulation. Sophisticated sequencing and other assay methods are now available to determine the genomic makeup (sometimes called “liquid biopsy”), which may obviate need for tumor biopsy in the future. However, circulating tumor cells are undetectable generally with stage I cancer or precancer. Tumor cells can also be recovered from stool but primarily with left-sided CRC and less with more proximal lesions.
Proteins
Classical protein markers, such as carcinoembryonic antigen, carbohydrate antigen 19-9, and alpha-1-antitrypsin, have been helpful for prognosis and surveillance in colorectal, pancreatic, and liver cancers, respectively, but have failed in early detection. Although proteins have the attractive features of test simplicity and low cost, many lack sufficient specificity or stability when assayed from distant media. Continued discovery efforts may yield discriminant marker candidates for future assays.
RNA
Various RNA species may be overexpressed or underexpressed in neoplasms and potentially serve as early detection markers. An advantage over DNA-based markers is that there are several thousand RNA copies per cell, which could translate into greater sensitivity and a requisite for smaller sample size. A challenge with many RNA markers has been nonspecificity and nonreproducibility across studies. Short RNA subtypes, such as microRNAs (miRNAs), may be more stable in stool or in blood.
DNA
The most studied class of tumor markers has been acquired DNA alterations, both genetic (eg, mutations, translocations) and epigenetic (eg, aberrant methylation). Important advantages of DNA markers include their distinguishing tumor-specific structural changes (rather than functional overexpression or underexpression, as occurs with protein or RNA) and relatively better stability. A technical challenge in targeting acquired genetic changes relates to the large assay capacity required to achieve broad diagnostic coverage, often involving interrogation of thousands of potential mutation sites across multiple genes; continued progress in high-speed sequencing methods may allow a practical solution someday. In contrast, aberrant methylation typically occurs at the gene promoter region or other predictable single sites within a gene, and a small panel of methylated DNA markers may provide broad coverage. Methylated DNA markers with site specificity have also been identified, a clinically relevant property that may allow the prediction of tumor location.
A glimpse at emergent and experimental new molecular tools for gastrointestinal cancer screening
New molecular tools may allow the accurate detection of both cancer and precancer at multiple GI sites, not only to create screening opportunities but to extend the diagnostic capabilities of the endoscopist as well.
Esophageal Cancer
Esophageal cancer is the sixth leading cause of cancer mortality worldwide. Esophageal adenocarcinoma predominates in Western countries and squamous cell carcinoma in Eastern countries. Incidence rates of both are increasing. Both cancer types are highly lethal when presenting symptomatically, readily curable when detected at presymptomatic early stages, and preceded by recognizable precursor lesions. Early studies have explored the feasibility of testing DNA markers in blood, saliva, and stool for the screen detection and prognostic assessment of esophageal cancer.
Sponge-on-string device
A simple, ingestible sponge-on-string device (SOS) has emerged as a potential minimally invasive, accurate, and low-cost approach to the screen detection of Barrett’s esophagus ( Fig. 2 ). Barrett’s esophagus, the metaplastic precursor to adenocarcinoma, may be present in more than 5% of the US population ; current approaches to its detection and surveillance are endoscopic. The feasibility of SOS has been established when linked with an immune assay for trefoil factor 3 (a marker of intestinal metaplasia) on the recovered cytologic sample. In a case-control study, this approach yielded a sensitivity for Barrett’s of 79.9% at a specificity of 92.4%; detection increased in proportion to Barrett’s length. Furthermore, modeling suggests that SOS is cost effective. Alternative molecular markers may have merit with SOS, because preliminary data using novel methylated DNA markers detected 100% of Barrett’s cases at 100% specificity (Iyer DDW 2016 abstract) . By either SOS sampling or endoscopic brushing, use of molecular markers may also complement biopsy for detection of dysplasia during surveillance. In a pilot study with a panel of methylated DNA markers applied to esophageal brushings, detection of Barrett’s low- and high-grade dysplasias was 71% to 78% and of small stage I cancer was 100%.
SOS may also have value in screening squamous cell carcinoma, a disease most prevalent in underserved Eastern populations. In a pilot study using SOS in Northern Iran, cytologic examination with p53 staining remarkably detected 100% of high-grade dysplasias at 100% specificity.
Gastric Cancer
Gastric cancer, the third most common cancer killer worldwide, is often curable with pre-symptomatic early-stage detection but remains unscreened in most regions. Thus, there is a compelling rationale for noninvasive molecular screening. To date, no validated screening tests have been brought to practice. Investigators have explored miRNA and methylated DNA markers in blood, and pilot DNA marker testing has suggested feasibility in stool as well. For example, a recent study assessing the discrimination of a miRNA panel in plasma found moderate discrimination for stage I cancer with areas under the receiver operating characteristics curve of 0.99 and 0.081 in training and test sets, respectively. Advances in whole methylome discovery have yielded highly accurate marker candidates with 100% sensitivity at 95% specificity (Anderson ACG 2015 abstract) . Technical optimization and further rigorous clinical testing are needed.
Gastric lavage
In a recent feasibility study, assay of methylated DNA markers in simple gastric washings detected 90% of gastric cancer at 96% specificity. Further investigation of patient acceptance and accuracy with this approach are needed.
Pancreatic Cancer
Most patients with pancreatic cancer present symptomatically at late stage. Its abysmal 5-year survival and alarming prevalence trend highlight the urgent need for improved early detection. Also, molecular tools may have adjunctive value for interrogation of the growing number of pancreatic cystic lesions incidentally found on abdominal imaging.
Pancreatic juice
Pancreatic juice can be collected readily from the duodenum by standard upper endoscopy after stimulation of secretin. In a recent pilot study, an assay of novel methylated DNA markers from pancreatic juice accurately detected pancreatic cancer with an area under the receiver operating characteristics curve of 0.92 ; distributions of a single marker, CD1D, in controls and cases with cancer are shown ( Fig. 3 ). Optimization and further evaluation in larger studies are indicated.
Cyst fluid
Management of pancreatic cysts represents a management conundrum, because most lesions are benign and structural features have proven inaccurate in predicting the presence of cancer or high-grade dysplasia. Various molecular markers have been explored to improve the detection of advanced dysplasia in cysts. A recent retrospective, multicenter study evaluated the combination of clinical features and DNA markers assayed from cyst fluid to detect lesions with mucinous features or dysplasia; cysts were accurately classified with a sensitivity of 90% to 100% and a specificity of 92% to 98%. Others have explored miRNA markers for this diagnostic application. Candidate methylated DNA markers have been identified able to discriminate high grade from low grade dysplasia in cysts with an area under the receiver operating characteristics curve of 0.91 [Majumder DDW 2016 abstract] ; applied studies are under way.
Blood
Blood testing may play roles in both staging and screening. For staging pancreatic cancer, blood taken via endoscopic guidance from the portal vein seems to yield more tumor cells than from matched peripheral blood. Circulating epithelial stem cells may also be present in blood with pancreatic cancer and even with premalignant cysts, and may be a source for molecular interrogation. In a recent study, investigators found that miRNA panels differentiated pancreatic cancer cases from healthy and chronic pancreatitis controls using peripheral whole blood with areas under the curve ranging from 0.86 to 0.93; discrimination was superior to that of carbohydrate antigen 19-9. Even higher discrimination for pancreatic cancer has been reported by serum assay of glypican-1 from circulating exosomes. Further studies are needed to corroborate these interesting findings.
Stool
Exploratory studies using nonoptimized markers and analytical methods have shown that pancreatic cancer can be detected by assay of exfoliated DNA markers in stool. Further development has the potential to increase the detection rates with this noninvasive approach.
Hepatobiliary Cancers
Worldwide, hepatoma represents the second major cancer killer. Although cholangiocarcinoma constitutes only 2% of malignancies, it is the second most common primary hepatobiliary cancer. There is a trend for rising incidence in the United States with both of these cancers.
Stricture brushing
The diagnostic evaluation of biliary strictures with conventional cytology obtained by endoscopic brushing has proved to be insensitive for detecting malignancy. In a recent study on biliary stricture brushings, complementary use of fluorescent in situ hybridization improved cancer detection to 65% compared with only 18% by cytology alone. Further improvements in diagnostic accuracy are needed. In a preliminary report, investigators assayed novel methylated DNA markers on archival brushings of biliary strictures and found a sensitivity of 100% at 90% specificity.
Cancer screening
Genome-wide methylation profiles have been described for hepatobiliary cancers at the tissue level and have variably been explored in distant biological media. Methylated DNA markers for hepatoma detection have been assessed in plasma, serum, and peripheral blood mononuclear cells with promising early results that suggest superiority over alpha fetoprotein levels. Candidate miRNA markers for hepatoma have been studied in plasma and serum. Pilot studies on molecular detection of hepatobiliary neoplasms have been reported using stool and urine.
Colorectal Cancer
CRC is currently the second most common cause of cancer death in the United States. Although screening reduces CRC mortality, conventional tools are variably accurate, associated with suboptimal compliance, and are not uniformly accessible. Innovative molecular approaches in stool or blood offer the promise of improved compliance and accessibility because of their noninvasive nature. Advances in molecular stool testing have recently led to a new commercial test with improved detection accuracy over fecal blood tests.
Multitarget stool DNA test
Approved by both the Food and Drug Administration and the Centers for Medicare and Medicaid Services in 2014 as the first molecular screening test for CRC, the new multitarget stool DNA test (MT-sDNA) was included in the 2015 American Cancer Society CRC screening guidelines and is now available for patient use (Cologuard, Exact Sciences, Madison, WI). A driver for the development of MT-sDNA was to improve participation rates based on its user-friendly features and ready access by mail. Early indicators seem to demonstrate success in this regard, because the test’s manufacturer has publicly reported that 42% of the first 100,000 users disclosed on a questionnaire that this was their first CRC screen.
MT-sDNA targets a panel of exfoliated DNA markers (mutant KRAS plus 3 informative methylated genes, BMP3 and NDRG4 ) and hemoglobin. In an initial multicenter study using a prototype MT-sDNA, it was established that detection rates for CRC and advanced adenoma were not affected by lesion site. Cutoffs for the optimized and automated MT-sDNA assay were established in a large case-control that which demonstrated sensitivities for CRC of 98% and for adenomas greater than 1 cm of 60% (72% for adenomas >2 cm, 83% for those >3 cm) at 90% specificity. MT-sDNA performance was then validated in a 10,000-patient multicenter study from the screen setting using colonoscopy as the reference standard. MT-sDNA detected 92% of CRC (94% for stages I and II) and 42% of advanced adenomas (66% for adenomas ≥2 cm) at a specificity of 90% (based on those with normal colonoscopy). For all lesion groups, detection rates by MT-sDNA exceeded those of fecal immunochemical test for occult blood (fecal immunochemical testing), although fecal immunochemical testing exhibited higher point specificity at 95%. A second cross-sectional screening study recently completed in Alaska Native people, who have among the world’s highest CRC incidence rates, showed MT-sDNA performance outcomes remarkably similar to those in the larger multicenter study ( Fig. 4 ).







