CHAPTER 102 Enteric Microbiota and Small Intestinal Bacterial Overgrowth
COMPOSITION AND MOLECULAR ANALYSIS OF THE ENTERIC MICROBIOTA
Most human enteric bacteria cannot be cultured, because of a lack of truly selective growth media. Nonetheless, molecular profiling has shown that whereas the microbiota appear distinct in different persons, the composition of each person’s microbiota is relatively stable after infant weaning and throughout adulthood. Evidence from studies of twins suggests that the individuality of human microflora may be genetically controlled,1 but environmental variables including diet and sanitation appear to have profound effects on early intestinal colonization with bacteria.2,3 In adulthood, dietary fluctuations appear to induce changes in bacterial enzymes and metabolic activity rather than changes in the relative populations of the microflora.2,4,5
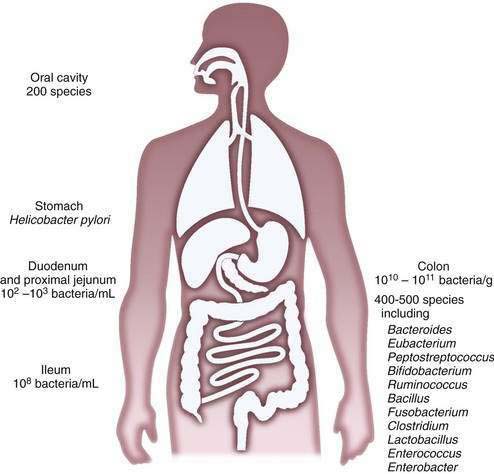
The composition of the microflora varies quantitatively and qualitatively over the longitudinal and the cross-sectional axes of the alimentary tract. Beyond the oral cavity, which harbors approximately 200 different bacterial species, the size and diversity of the microflora increase distally along the digestive tract (Fig. 102-1). Gastric acid restricts bacterial numbers within the stomach to fewer than 103 colony-forming units (CFU)/mL. The gradient in bacterial density is greatest across the ileocecal valve, with approximately 108 bacteria per gram of ileal contents and up to 1012 bacteria per gram of colonic contents, comprising more than 1000 different bacterial species.2,4–6 More than 99% of the culturable bacteria in the ileum and the colon are obligate anaerobes, but the composition of the flora at the mucosal surface differs from that within the lumen; ratios of anaerobes to aerobes are lower at mucosal surfaces. Culture-independent methods, such as the various molecular approaches described later, suggest that mucosa-associated bacteria differ from those recovered from feces, thus supporting the idea that host-related factors have a role in determining the enteric microflora7 and implying that bacterial aspirates from the lumen may be an incomplete reflection of mucosa-associated bacteria.
The microbiota of the proximal small intestine consist predominantly of Gram-positive facultative bacteria—bacteria that can survive under aerobic or anaerobic conditions—although enterobacteria and Bacteroides species also may be present. Peristalsis is the principal factor restricting bacterial numbers in the small intestine. In the distal small intestine, the composition of the microflora resembles that of the colon, with a preponderance of Gram-negative anaerobes. The most prominently represented genera in the distal bowel include Bacteroides, Clostridium, Lactobacillus, Fusobacterium, Bifidobacterium, Eubacterium, Peptococcus, and Escherichia species.2,6
Detailed analysis of the enteric microflora has been confounded by the limitations of traditional culture-dependent microbiology.8 First, obtaining representative material from different niches within the intestine is problematic; because most of the indigenous microflora are obligate anaerobes, major methodologic difficulties are encountered with sampling, contamination, transport, and storage. Second, the lack of truly selective growth media precludes culture of most components of the microflora. In this respect, it is noteworthy that culture of H. pylori and C. difficile was accomplished only within the past two to three decades. This difficulty has led to a shift in emphasis from conventional bacterial phenotyping toward genotyping and molecular approaches to study the unculturables.4,9,10
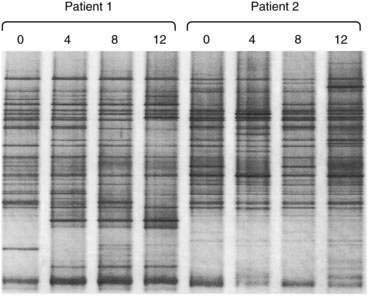
For rapid profiling of the dominant culturable and nonculturable organisms within a complex ecosystem such as that in the intestinal tract, 16S rRNA can be amplified by polymerase chain reaction (PCR) with universal primers spanning conserved and variable regions. The mixture of hypervariable RNA fragments can then be separated by a chemical denaturing gradient or a temperature gradient gel electrophoresis (DGGE and TGGE). Complete denaturation of the RNA fragments is prevented by incorporating a GC-rich 5′ end to one of the primers (a GC clamp).11 Variations in migration distance through the denaturing gradients reflect the diversity of 16S species in the sample (Fig. 102-2). Theoretically, the technique is semiquantitative, because the more dominant the organism, the more abundant the specific PCR product. The specific PCR product can be cut from the gel and further amplified, cloned, and sequenced to identify individual bacterial strains without requiring a conventional culture step. Further refinements of the technique can be achieved by using species-specific PCR primers.
Other molecular techniques for analysis of specific bacterial species now are possible because of the increasing availability of genomic sequence data for the major components of the bacterial flora. These techniques include fluorescence in situ hybridization (FISH) flow cytometry (FISH-flow) and bacterial DNA microarrays. In disorders such as inflammatory bowel disease, immunologic reactivity against components of the microflora has been used to identify microbes that may be etiologic in the disease. Marker antibodies generated by hybridoma or phage-display technology have been used as reagents to identify microbial antigens. For example, antineutrophil cytoplasmic antibody (pANCA), which is associated with ulcerative colitis, has been used to identify colonic bacteria expressing a pANCA-related epitope.12,13 In addition, candidate microbes can be identified by the presence of unique bacterial nucleic acid sequences associated with a particular lesion or disease location, by subtractive cloning using genomic representational difference analysis. With this technique, a sequence representing a bacterial transcription factor from an apparent commensal organism, Pseudomonas fluorescens,14–16 was found in lesions of Crohn’s disease but not in adjacent nonlesional mucosa. Serologic expression cloning also has been used to identify bacterial flagellin as a dominant antigen in Crohn’s disease.17
The new science of metagenomics—the sequencing of genes from whole microbial environments at once—promises to address many of the unresolved questions about the microbiota. The microbiota comprise a repository of genetic information (microbiome) that greatly exceeds that of the host genome. By combining metagenomics with bioinformatics, biochemistry, and traditional bioassays, new insights into the metabolic capacity of the human intestinal microbiota can be achieved.4 Major consortia (including the human microbiome project4) around the globe are under way using metagenomics as a tool for bioprospecting the intestinal microbiota in health and disease.
INTERACTIONS BETWEEN HOST AND MICROBES
The microbiota exert both positive and negative regulatory effects on the development and function of the intestine. These complex influences first were shown in comparative studies of germ-free and conventionally colonized animals. A sterile intestine is associated with reductions in mucosal cell turnover, digestive enzyme activity, cytokine production, lymphoid tissue, lamina propria cellularity, vascularity, muscle wall thickness, and motility, but with an increase in enterochromaffin cell area.18 The molecular events underpinning this regulatory signaling from the lumen currently are being explored using modern techniques such as laser capture microdissection and gene array analysis; such studies promise to reveal new molecular targets to be exploited for the design of novel therapeutics.19,20 Thus, for example, when applied to animals colonized with only a single bacterial strain, Bacteroides thetaiotaomicron, this combined approach has illustrated the impact of bacteria-derived signaling on the expression of host genes controlling mucosal barrier function, nutrient absorption, angiogenesis, and development of the enteric nervous system.
Incoming bacterial signals include secreted chemoattractants, such as the formylated peptide f-Met-Leu-Phe, cellular constituents such as lipopolysaccharide (LPS) and peptidoglycans, flagellin, and bacterial nucleic acids (i.e., CpG DNA). Detection of bacterial stimuli by the host and discrimination of pathogens from commensals are mediated in part by pattern recognition receptors such as Toll-like receptors (TLRs) that are present on epithelial and immune (dendritic) cells. In health, engagement of TLRs by ligands from the commensal microbiota appears to be required for mucosal homeostasis.21,22 Thus, not only are bacterial signals required for optimal mucosal and immune development, but they also actually are required to maintain and condition the mucosa for responses to injury.22
The immune system mediates the sense of microbial danger and responses to injury. Although the primary lymphoid organs are developed at birth, mucosal immune functions require continual education and fine-tuning of cytokine balances and T-cell responses; this process is achieved by microbial colonization and sporadic mucosal infections. Without the microbiota, mucosal lymphoid tissue is rudimentary, and induction of mucosal immune responses and tolerance is suboptimal.23,24
With a surface area similar to that of a tennis court (approximately 400 m2) and only one cell layer separating the internal milieu from the lumen, the enteric mucosa is well adapted to immunologic sampling of the intraluminal microbial community. Sampling of the microbiota across the epithelial barrier is mediated by M cells, which deliver particulate and microbial antigens to underlying immune cells, and by mucosal dendritic cells, which appear to extend processes into the lumen between the surface enterocytes without disrupting tight junctions.25 It appears that intestinal dendritic cells can ingest and retain intact live commensal bacteria and then transit to the mesenteric lymph node, where immune responses to commensals are induced locally.26 Thus, the mesenteric lymph node acts as a gatekeeper, preventing access of commensal bacteria to the internal milieu and protecting the host from harmful systemic immune reactivity. The immunosensory function of dendritic cells is facilitated by their plasticity and versatility of responses,27 depending on whether they are presented with commensals or pathogens; moreover, they appear to exhibit tissue-specific specialization in the intestine.23,24
In addition to specific immune responses to enteric bacteria, the surface epithelial cells serve a sensory function to detect microbial danger by producing chemokines that activate the host immune response and recruit it to any breach in the mucosal barrier caused by pathogenic infection.28
Transduction of bacterial signals into host immune responses after engagement of TLRs may proceed along more than one molecular pathway. The transcription factor nuclear factor-κB (NF-κB) is the pivotal regulator of epithelial responses to invasive pathogens, but nonpathogenic bacteria can attenuate inflammatory responses by delaying the degradation of IκB, which is counter-regulatory to NF-κB.29 Other signal transduction pathways are likely to emerge to account for the anti-inflammatory effects of probiotics and other commensal organisms such as Bacteroides thetaiotaomicron. This anaerobic commensal can antagonize the proinflammatory effects of NF-κB within the epithelial cell by enhancing the nuclear export of its transcriptionally active subunit (RelA) in a peroxisome proliferator-activated receptor-γ (PPAR-γ)–dependent manner.30
METABOLIC ACTIVITY OF THE MICROBIOTA
The enteric microbiota are tantamount to a hidden metabolic organ (Table 102-1). Although our understanding of indigenous bacterial metabolites is still superficial, coevolution with this living inner mass of bacteria has several apparent benefits for the host. In addition to the production of regulatory signals for mucosal homeostasis as discussed earlier, the microbiota exhibit important metabolic properties not possessed by the host. These include biotransformation of bile acids; degradation of oxalate; breakdown of otherwise indigestible dietary components, such as plant polysaccharides; and production of short-chain fatty acids, a major energy source for colonic epithelium, from fermentable carbohydrates. Other activities include synthesis of biotin, folate, and vitamin K.2,20 Clinicians also have exploited enteric bacterial enzymes such as azoreductase to convert prodrugs such as sulfasalazine to active drug metabolites (e.g., aminosalicylate). Other examples of bacterial action on drug bioavailability include the metabolism of l-dopa to dopamine and degradation of digoxin. Not all of the metabolic changes induced by the enteric microbiota are beneficial to the host, however, and although bacteria probably degrade some carcinogens, they also might promote the production of carcinogens from dietary procarcinogens.31
Table 102-1 Examples of Metabolic Activities of Intestinal Microbiota
A striking example of the importance of bacterial metabolism is exemplified by the regulatory effect that the enteric microbiota exert on fat storage.32 It has long been known that germ-free animals need a significantly greater caloric intake to sustain a body weight similar to that of normal colonized animals. Thus, the normal host-microbiota relationship has nutritional benefit, in contrast with the negative nutritional effect associated with bacterial overgrowth syndromes. Elegant studies with germ-free mice have shown that upon colonization, body weight increases despite a reduced caloric intake. The bacteria in the microbiota colonizing the intestine promote storage of dietary calories in fat by increasing absorption of monosaccharides and suppressing epithelial-derived fasting-induced adipocyte factor (FIAF).32 Thus, the composition and activity of the intestinal microbiota should be considered as a diet-influenced variable that can influence susceptibility to obesity.
One of the outcomes of bacterial metabolic activity is gas production. Of the five gases—N2, O2, CO2, H2, CH4—that constitute 99% of flatus, the latter three are produced by the enteric bacteria, and bacteria are the sole source of hydrogen and methane in the intestine. Hydrogen production by bacterial action on carbohydrates, and to lesser extent on protein, normally occurs in the colon. In patients with small intestinal bacterial overgrowth, however, the small intestine also becomes a site of H2 production. Bacterial methanogens occur in the colon and produce methane from H2 and CO2, with significantly detectable excretion in approximately 30% of humans.33–37 The principal gases produced are odorless, but bacterial metabolism also is responsible for producing various trace and odiferous gases in flatus such as hydrogen disulfide.38,39 Qualitative and quantitative variability in gas production with diet illustrates the fluctuations in bacterial metabolic activity despite the apparent stability of the microbiota in adulthood.
SMALL INTESTINAL BACTERIAL OVERGROWTH
Small intestinal bacterial overgrowth (SIBO) is characterized by malabsorption and overgrowth of bacteria in the small intestine. The syndrome often is referred to as blind loop syndrome because of recognition of the disorder in patients with predisposing anatomic abnormalities. Other terms that have been used to describe the disorder include stagnant loop syndrome, contaminated small bowel, small intestinal colonization, and small bowel stasis. In 1939, Barker and Hummel40 reported macrocytic anemia in association with intestinal strictures and anastomoses and postulated that the anemia was secondary to bacterial overgrowth, or “putrefaction.” SIBO is not confined to humans and is well recognized in dogs.41 The syndrome is associated with a variety of anatomic disturbances, such as blind loops,42 and motility disorders, such as scleroderma,43 but it can occur in the absence of any specific predisposing factor. It is likely that the condition is underdiagnosed, particularly in the elderly.44
Patients with SIBO do not necessarily present with a florid malabsorption syndrome, and symptoms may be minor and nonspecific. Considerable debate has concerned the relationship between irritable bowel syndrome (IBS) and SIBO (see Chapter 118).45 Bacterial overgrowth has been documented in asymptomatic elderly persons in the community,46,47 in whom it is debatable whether the phenomenon is of any significance48; in the absence of malabsorption or related symptoms, such overgrowth probably should not be considered to represent true SIBO. Asymptomatic SIBO may be termed simple colonization and probably results from achlorhydria and abnormal fasting intestinal motility (see later).
The diagnosis of SIBO usually is made by noninvasive breath testing,49 even though studies on the accuracy of these tests report very variable results. For this reason, culture of a small intestinal aspirate must be regarded as the diagnostic gold standard. Unfortunately, much of the published literature on SIBO is based on breath tests, rather than on culture, and the findings must be interpreted with caution.
ETIOLOGY AND PREDISPOSING FACTORS
The upper small intestine is an environment of relatively low bacterial counts because of the combined effects of gastric acid and peristalsis. Bacterial counts in aspirates from the normal upper small intestine generally are less than 1000/mL. Pathophysiology and predisposing conditions are listed in Table 102-2.
Table 102-2 Pathophysiology and Some Conditions Associated with Small Intestinal Bacterial Overgrowth
| PATHOPHYSIOLOGY | CONDITION |
|---|---|
| Anatomic abnormalities | |
| Motility disorders | |
| Reduced gastric acid secretion | |
| Abnormal connection between colon and proximal bowel | |
| Various mechanisms |
Anatomic Abnormalities
A variety of anatomic abnormalities, including iatrogenic and disease-related abnormalities, lead to stagnation of small intestinal contents, resulting in bacterial overgrowth. The bacteria in SIBO are similar to those found in the normal colon, and certain organisms are common. Common aerobic organisms include Escherichia coli and Streptococcus, Staphylococcus, Micrococcus, Klebsiella, and Proteus species. Common anaerobic species include Lactobacillus, Bacteroides, Clostridium, Veillonella, Fusobacterium, and Peptostreptococcus.50 The classic anatomic cause of SIBO is a blind loop resulting from abdominal surgery, such as Billroth II partial gastrectomy; other anatomic abnormalities that can result in SIBO include intestinal strictures and small bowel diverticulosis.
Motility Disorders
Disorders affecting small intestinal peristalsis, such as scleroderma,43 diabetes mellitus,51 and chronic idiopathic intestinal pseudo-obstruction52 constitute the next most common cause of SIBO after anatomic abnormalities.
SIBO is well recognized in scleroderma and occurs mainly in patients with small intestinal involvement72 who have limited cutaneous systemic sclerosis. Diarrhea is the most important symptom. The somatostatin analog octreotide is effective in the management of SIBO associated with scleroderma.73
Although small intestinal dysmotility is thought to be the main predisposing factor in diabetes, SIBO in diabetics is not especially associated with autonomic neuropathy.74 Treatment of SIBO in diabetics improves orocecal transit time.75
Fistula or Ileocecal Valve Resection
The ileocecal valve prevents reflux of colonic bacteria into the small intestine, and resection of the valve or development of fistulas between the colon and upper gastrointestinal tract can lead to reflux of colonic contents into the small intestine, with ensuing bacterial overgrowth.53,54
Reduced Gastric Acid Secretion
Achlorhydria is known to be a predisposing factor for SIBO, and SIBO has been described in patients after vagotomy,55 in those with atrophic gastritis, and in those taking acid suppressants.56–59 SIBO occurs more often in patients taking proton-pump inhibitors (PPIs) than in those taking histamine H2 receptor antagonists,58 but clinical malabsorption does not appear to occur in this situation.59
Aging
Advancing age seems to be an independent risk factor for SIBO, but it is not clear if overgrowth results from the aging process itself and age-related changes in intestinal motility or if it is a consequence of achlorhydria. Early studies in this area found that SIBO was a common (and commonly unrecognized) cause of malabsorption in the elderly44,60 and that many such patients did not have an obvious predisposing factor, such as a blind loop. More-recent studies have reported SIBO in asymptomatic elderly persons residing in the community. These patients, although asymptomatic, had lower weights and body mass indices (BMI) than expected, and treatment with antibiotics increased both weight and BMI.46,47 In contrast, a Japanese study reported SIBO (diagnosed by glucose hydrogen breath test) in 25.6% of disabled older adults but in none of the healthy older adults.61
Chronic Liver Disease
SIBO appears to be common in patients with chronic liver disease,62,63 is more common in patients with advanced (Child class C) liver disease,63 and may be an independent risk factor for spontaneous bacterial peritonitis,64 although this association is controversial.65 No association with any particular cause of chronic liver disease has been found,66 but SIBO does not occur in cirrhotic patients who do not have portal hypertension.67 The etiology of SIBO in patients with chronic liver disease is likely to be related to disturbances in gastrointestinal motility67 and possibly to the use of antacids,65 both of which can foster proliferation of bacteria. Small intestinal dysmotility is more severe in cirrhotic patients with a history of spontaneous bacterial peritonitis,64 and treatment of SIBO improves motility.68 Liver transplantation improves small bowel dysmotility in cirrhotic patients.68 Antibiotics and prokinetic agents are effective in reducing the SIBO associated with cirrhosis.69 SIBO in cirrhosis is associated with systemic endotoxemia.70 Oral conjugated bile acids reduce bacterial overgrowth and endotoxemia in cirrhotic rats, suggesting a contributory role for cholestasis in cirrhotic patients with SIBO.71
Other Causes
SIBO is present in many patients with celiac disease who have persistent symptoms despite their adherence to a gluten-free diet.76 It is not clear why this is so, but a motility disturbance seems the most likely explanation.
SIBO is common in Crohn’s disease, particularly in patients who have had previous intestinal resection, and orocecal transit time has been reported to be prolonged in Crohn’s patients with SIBO.77 Positive results on glucose hydrogen breath tests are particularly associated with the presence of a small bowel stricture.78
SIBO is common in chronic pancreatitis.79 SIBO in this setting may be caused by small bowel dysmotility resulting from chronic opioid use and achlorhydria. Furthermore, pancreatic juice may have an antibacterial effect, so its absence might allow enteric bacteria to proliferate more freely.80
SIBO in rheumatoid arthritis is associated with high disease activity and does not appear to be related to achlorhydria.81
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree