Bladder cancer is a heterogeneous disease characterized by complex networks of molecular alterations and gene expression. This review summarizes some of the recent genomic studies that have further advanced the understanding of the pathways driving bladder cancer, highlighting several important biomarkers and potential targeted therapeutic strategies that are now in clinical trials. In addition, noninvasive techniques to evaluate biomarkers in patients’ urine and serum for early detection and surveillance are discussed.
Key points
- •
Bladder cancer is a heterogeneous disease, and recent genomic studies have identified several potential therapeutic targets.
- •
Alterations in tyrosine kinase receptors, intracellular signaling pathways, such as the PI3K/AKT/mTOR pathway, cell-cycle regulators, chromatin remodeling, and immune mediation, are significant in disease progression, and therapies targeting many of these alterations are currently in clinical trials.
- •
Novel noninvasive strategies are being developed, using identification of genomic, epigenetic, and proteomic markers, for early detection and surveillance in urine and serum.
Genetic and molecular biomarkers
Deciphering the molecular pathways of bladder cancer has accelerated the identification of prognostic and theranostic markers, allowed for the development of novel noninvasive early detection and surveillance strategies, and elucidated new targets of therapy in bladder cancer. Although widespread clinical adoption of novel biomarkers has been limited due to lack of validation by multi-institutional randomized prospective trials, recent genomic studies have spurred efforts to evaluate these therapies, and such trials are finally being launched.
Genetic and molecular biomarkers
Deciphering the molecular pathways of bladder cancer has accelerated the identification of prognostic and theranostic markers, allowed for the development of novel noninvasive early detection and surveillance strategies, and elucidated new targets of therapy in bladder cancer. Although widespread clinical adoption of novel biomarkers has been limited due to lack of validation by multi-institutional randomized prospective trials, recent genomic studies have spurred efforts to evaluate these therapies, and such trials are finally being launched.
Genomics of bladder cancer
Recent genomic studies have validated and expanded on previously identified genetic pathways of bladder cancer development and have unmasked additional crucial driver genetic alterations. Although earlier array-based gene expression studies highlighted differentially expressed genetic signatures capable of predicting recurrence and progression, recent integrated genomic and protein analysis studies have better defined clinically relevant molecular subtypes of bladder cancer. By integrating genomic data from aCGH, gene expression arrays, targeted mutation sequencing analysis, and protein analyses, Lindgren and colleagues brought to light 2 main genomic molecular circuits in urothelial carcinoma: the first characterized by FGFR3 alterations, overexpression of CCND1 , and deletions in 9q and CDKN2A ; and the second by E2F3 amplifications, RB1 and PTEN deletions, gains of 5p, and overexpression of CDKN2A (p16). Alterations in TP53/ MDM2 were demonstrated in advanced tumors in both groups. Lindgren and colleagues first recognized the significantly worse prognosis associated with a gene expression profile of a keratinized/squamous phenotype; this molecular subtype was further validated by Choi and colleagues ( Fig. 1 ). Termed basallike and characterized by p63 activation, squamous differentiation, positive CK5/6, epidermal growth factor receptor (EGFR), and cluster of differentiation (CD)44 expression and lack of cytokeratin (CK)20, this subtype is clinically aggressive but potentially sensitive to neoadjuvant chemotherapy. Choi and colleagues also characterized a luminal subtype typically enriched for activating FGFR3 mutations, active estrogen receptor pathway, and ERBB2 and PPARγ expression profile, and a third subtype, characterized by wild-type TP53 gene expression and strongly associated with resistance to neoadjuvant methotrexate, vinblastine, adriamycin/doxorubicin, and cisplatin (MVAC) therapy. Interestingly, upon resistance to chemotherapy, tumors from the basallike and luminal subtypes also displayed the TP53 wild-type expression.
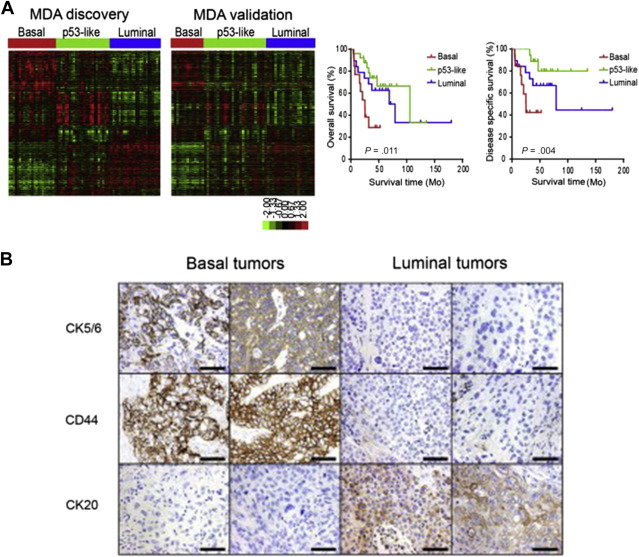
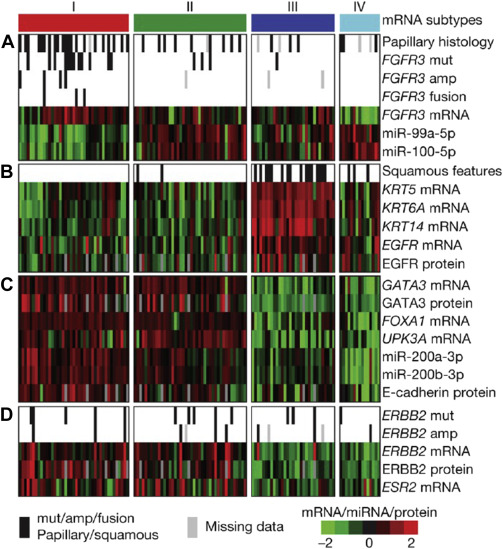
Finally, The Cancer Genome Atlas (TCGA) project’s comprehensive molecular characterization of bladder cancer provided a genomic analysis of 131 high-grade muscle-invasive bladder cancers (MI-BC), which revealed a staggering 302 mutations, 204 segmental copy number alterations (CNA), and 22 rearrangements on average per tumor. Recurrent “driver” mutations in 32 genes were found, which include genes involved in cell-cycle regulation, chromatin regulation, kinase signaling pathways, and 9 additional genes not previously shown to have recurrent mutational pattern in other tumors. Based on integration of mRNA/miRNA sequencing data and protein expression analysis, 4 major expression clusters were identified ( Fig. 2 ). Among them, papillarylike cluster (cluster I), enriched for FGFR3 gene alterations, together with cluster II share expression of luminal urothelial differentiations markers (activated expression of ER, GATA3, Uroplakin, and ERBB2) and cluster III basal/squamouslike, as in the Choi and colleagues study, characterized by CK5/6 and EGFR expression ( Table 1 ). When compared with the TCGA profiles of several other tumor types, bladder cancer clusters I and II are very similar to luminal A breast cancer subtype, whereas cluster III is similar to basallike breast cancer, and squamous cell carcinoma of the head and neck and lung.
| Subtype/Cluster | Molecular Characteristics | Histology (General) | Noteworthy Clinical Features |
|---|---|---|---|
| Non-muscle-invasive bladder cancer | |||
| Papillary | FGFR3 mutations 9q deletions CDKN2A deletions (9p21) | Papillary | Low grade: Recurrence risk ∼50% Risk of progression (∼5–10%) High grade: Recurrence risk >50% Risk of progression (15%–40%) |
| Muscle-invasive bladder cancer | |||
| Subtype/TCGA cluster based on RNA sequencing | Molecular characteristics | Histology (general) | Noteworthy clinical features |
| Luminal (I) (similar to luminal A breast cancer subtype) | FGFR3 mutations FGFR3-TACC3 fusions CDKN2A deletions (9p21) HER2 expression ESR2 expression miR-200 family (EMT) | Papillary-like | Chemosensitive |
| P53-like (II) (similar to luminal A breast cancer subtype) | HER2 expression ESR2 expression miR-200 family (EMT) | Chemoresistant | |
| Basal/squamous-like (III) (similar to basal-like breast cancer, squamous cell carcinoma of head and neck and lung) | Express cytokeratins (KRT14, KRT5) CCND1 amplification | Squamous-like; sarcomatoid | Chemosensitive |
| Claudin-low (IV) | — | — | Uncertain |
| Special variant histology of note | |||
| Molecular characteristics | — | Noteworthy clinical features | |
| Micropapillary | ERBB2 mutations and amplification | — | Aggressive disease |
| Plasmacytoid | Gains: 11q, 17q, 17p, 20q; losses on 4q and 6q; CCND1 and CDH1 deletions (25087089) | — | Aggressive disease |
Biomarkers and pathway targets
Receptor Tyrosine Kinases and Cell-Cycle Markers
Numerous studies suggest a prognostic value for receptor tyrosine kinases, such as FGFR3 , EGFR, ERBB2, and ERBB3 in bladder cancer ; for instance, FGFR3 mutations correlate with a favorable outcome. An elevated tumor proliferation index, measured by ki67 or Monoclonal Anti-Human Ki-67 Antigen, Clone MIB-1 (MIB-1) immunohistochemistry, consistently predicts a worse outcome in bladder cancer. A molecular grade (mG), initially proposed by Van Rhijn and colleagues, combining FGFR3 gene mutation status and MIB-1 index, was found to be superior to the NBI-BC risk calculator of the European Organization for Research and Treatment of Cancer. mG independently predicts disease-specific survival (DSS), and when added to the multivariable model for progression, increases the predictive accuracy to 81.7%. Proliferation index is also prognostic in MI-BC ; a large cystectomy cohort from the bladder consortium multi-institutional trial confirmed the role of Ki67 in predicting progression-free survival (PFS) and DSS. In addition, a synergistic prognostic role for combining p53 expression status with other cell-cycle control elements, such as pRb, cyclin E1, p21, and p27, has been repeatedly shown in non-muscle-invasive bladder cancer (NMI-BC) as well as MI-BC ( Fig. 3 ).
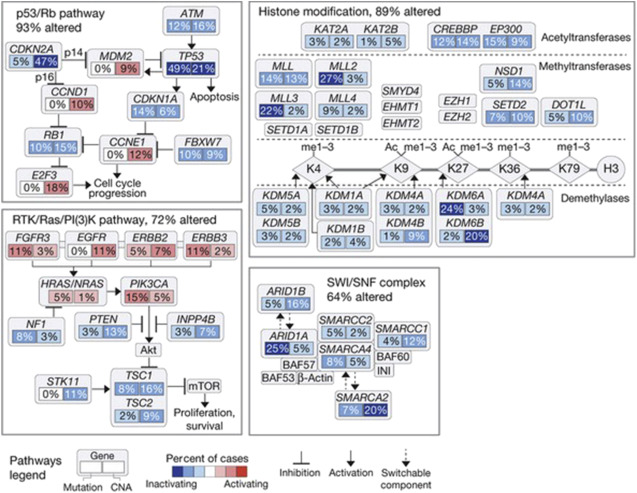
Epigenetic Markers
Both gene methylation and miRNAs have been successfully analyzed in patients with bladder cancer, demonstrating their potential utility as tools for early detection and prognosis. Epigenetic profiles, specifically, promoter methylation of RASSF1A , DAPK , CDH1 (encodes for E-cadherin), TNFSR25 , EDNRB , and APC genes, have been shown to predict disease progression and possibly death from disease independent of tumor stage.
Monitoring/detection methods
With the goal of early detection and improved surveillance, less invasive techniques are being investigated in order to develop the “liquid biopsy” to measure biomarkers in either blood or urine. Urovysion, which is US Food and Drug Administration (FDA) approved for surveillance as well as screening and early detection in high-risk patients with hematuria, exploits recurrent chromosomal alterations (gains of chromosomes 3q, 7p, and 17q, and 9p21 deletions [ CDKN2A /p16 locus]) in a urine-based multitarget interphase fluorescence in situ hybridization (FISH) assay. When used as a reflex test in combination with routine urine cytology, a sensitivity of 69% to 87% and specificity of 89% to 96% are achieved. In addition, up to two-thirds of patients with positive FISH but clinically absent disease have been shown to develop cancer within 29 months, suggesting a lead time value for molecular early detection.
More recently, urine-based assays for FGFR3 mutation, given the high prevalence in NMI-BC, and TERT promotor mutations, either as a single marker or in combination with other alterations, are being evaluated. Table 2 further elucidates these and some other biomarker assays that are currently available and are in development for this purpose.
| Product | Methodology/Target | Sensitivity/Specificity | Reference |
|---|---|---|---|
| Commercially available products (representative) | |||
| Multicolor FISH (FDA approved; Urovysion; Abbott Molecular, Abbott Park, IL, USA) | FISH for 4 chromosomal markers: gains of 3, 7, and 17 and loss of 9p21 | Sensitivity of 69%–87% Specificity of 89%–96% when combined with urine cytology | |
| ImmunoCyt (FDA approved; Scimedx, Denville, NJ, USA) | Detects 3 different proteins: M344, 19A211, and LDQ10 on the surface of voided cells (fluorescence immunohistochemistry) | Sensitivity 50%–100% for early stage disease Overall sensitivity 62% Overall specificity 79% | |
| Developing methods | |||
| FGFR3 mutation in combination with methylation markers (PCR based) | Sensitivity 62% (urine) Specificity 100% | |
| TERT promoter mutation (PCR based) | Variable according to clinical indication, tumor grade and methodology | ||
| Urine cytology | Sensitivity 48% Specificity 86% (Sensitivity for LG, 16%; HG, 84%) | ||
| DNA-based; RNA-based; DNA methylation; miRNA; long noncoding RNA | — | |
| Upregulation of calgranulin A (S100A8), calgranulin B (S100A9), calcium binding protein (S100A4), carbonic anhydrase I, and downregulation of calcium-dependent phospholipid-binding protein (annexin V) (MALDI-TOF-MS) | Sensitivity 80% (serum) Specificity 81% | |
| An 8-biomarker panel: IL-8, MMP-9, PAI-1, VEGF, ANG, CA-9, APOE, MMP-10 (ELISA) | Sensitivity 92% (urine) Specificity 97% | ||
Clinical trials
The TCGA and preceding comprehensive genomic studies have identified a spectrum of therapeutic targets that are present in the majority (>70%) of MI-BC. Clinical trials investigating such targeted therapeutic strategies along with identification and validation of predictive markers that correlate with response to therapy are underway. Key targets include members of the PI3KCA/AKT/mTOR , RTK/MAPK (including EGFR, FGFR3, and ERBB2 ), and estrogen receptor ( ER ) pathways, immune response check point modulators, and chromatin regulation and remodeling targets ( Table 3 ).
| Altered Gene/Protein | Drug Category | Drug Examples | Clinical Trial Examples (Phase) |
|---|---|---|---|
| RTK/RAS pathway | |||
| KRAS, HRAS, NRAS, BRAF, EGFR | MEK inhibitor | Selumetinib | |
| EGFR inhibitor or antibody | Cetuximab; erlotinib | NCT00749892 (2) NCT00380029 (2) | |
| FGFR1, FGFR2, FGFR3 | pan-FGFR inhibitor | BGJ398; dovitinib | NCT01004224 (1) NCT01928459 (1) NCT01732107 (2) |
| ERBB2, ERBB3 | ERBB2 inhibitor | Trastuzumab; lapatinib a ; neratinib a ; DN24–02; T-DM1 | NCT02342587 (2) NCT01353222 (2) NCT01953926 (2) |
| VEGF | VEGF inhibitor | Bevacizumab; sorafenib; pazomanib | NCT01108055 (2) |
| MET | MET inhibitor | Cabozantinib (XL184) | NCT02496208 (1) |
| PI3K/AKT/mTOR pathway | |||
| PIK3CA, AKT1, AKT3, TSC1, TSC2, PTEN | AKT inhibitor | MK2206; AZD5363 | — |
| mTOR inhibitor | Everolimus; temsirolimus; AZD2014 | NCT01259063 (1) | |
| pan-PI3K inhibitor | BKM120 (buparlisib); BYL719 | NCT01470209 (1) (combined with everolimus) | |
| Cell-cycle regulation | |||
| AURKA, PLK1, CDK4, CCND1/CCND3, CDKN2A | Cdk4/6 inhibitor | LEE011; Palbociclib (PD-0332991) | NCT02187783 (2) NCT02334527 (2) |
| Chromatin remodeling/histone modification | |||
| ARID1A, MLL2, KDM6A, EP300 | Agents that bind to acetyl-lysine binding motifs (bromodomains) | BMS-986158; OTX015; TEN-010 | |
| Hormonal therapies | |||
| ESR2 upregulation (estrogen receptor) | Estrogen receptor modulator | Tamoxifen; raloxifene | |
| Immune modulators | |||
| CTLA4 | Anti-CTLA4 | Ipilimumab; tremelimumab | |
| PDL1 (CD274) PD1 | Anti-PDL1 Anti-PD1 | MPDL3280A; MED14736; Pembrolizumab (MK-3475) | NCT02450331 (3) NCT02108652 (2) NCT02302807 (3) NCT02335424 (2) NCT02256436 (3) |
| Stress response | |||
| HSP upregulation | HSP90 inhibitor HSP27 inhibitor | Ganetespib OGX-427 | NCT01780545 (2) |
| Cancer stem-cell expression | |||
| KRT14, KRT5 | — | — | NCT02027649 (NA) (KRT14 detection) |
| Basket trials | |||
| NCI-MATCH | — | — | Many |
| Signature trials | — | — | Many |
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree




