Fig. 17.1
The human orthologs identified from the rat proteins in perfusion-driven urine were compared with human kidney expression data (Kidney expr), the pooled human urine and urinary exosome proteome (UriANDexo), and the human plasma proteome (Plasma). The protein identifiers were standardized using the Ensembl Gene ID(s). The comparison was performed at the gene level [28]
Of the 1,278 genes, 982 were expressed in the kidney. These genes corresponded to 981 human orthologs. The 981 human orthologs with gene expression in the kidney were considered to be potential human kidney proteins in urine. Of the 981 human orthologs, 613 had been identified both in the urine (including urinary exosome) proteome and the plasma proteome; 240 had only been identified in the urine (including urinary exosome) proteome but not in the plasma proteome; 71 had only been identified in the plasma proteome but not in the urine (including urinary exosome) proteome; and 57 had not been identified in either the urine (including urinary exosome) proteome or the plasma proteome (Fig. 17.1).
There are a total of 128 human orthologs (57 plus 71) that were expressed in the kidney but were not present in normal urine (including the urinary exosome). They are potential biomarkers with zero background in pathological conditions. There are a total of 297 human orthologs (57 plus 240) that were expressed in the kidney but were not present in the plasma. They are likely not influenced by other normal organs, including the plasma and therefore have the potential to specifically reflect functional changes in the kidney. The 57 human orthologs could be sensitive markers because they were not present in normal urine or the urinary exosome and were not influenced by other normal organs, including plasma.
17.6 Comparing the Ranking of Human Kidney Origin Proteins in the Normal and Perfusion-Driven Urine
One large-scale dataset of the human normal urine proteome was collected from one team in our institution. They used the same MASCOT search engine as in this study. The Exponentially Modified Protein Abundance Index (emPAI), which offers approximate, label-free, relative quantitation of the proteins in a mixture based on protein coverage by peptide matches, has been incorporated into the MASCOT search engine [26]. Therefore, each identified urine protein had an emPAI value, which can be used to approximately estimate the absolute protein contents in urine.
Of the 981 human orthologs that were considered to be potential human kidney origin proteins in urine, 775 were identified in this normal human urine dataset. The emPAI values of these human orthologs were extracted from the normal human urine proteome, and these proteins were sorted from most to least abundant in the normal human urine. Proteins not identified in the human urine were at the end. The order of these human orthologs approximately represents their abundance in human urine under physiological conditions.
The 981 human orthologs were paired to rat proteins that were identified in both isolated rat kidney perfusion-driven urine samples. The rat proteins corresponding to human orthologs had an emPAI value when they were identified in the perfusion-driven urine, which can be used to approximately estimate the absolute protein content in the perfusion-driven urine. The rat proteins corresponding to the human orthologs were sorted according to their emPAI values in the two perfusion-driven samples from most to least abundant in the perfusion-driven urine. Due to the correspondence between rat proteins and their human orthologs, this resulted in the reordering of the 981 human orthologs. We assume that the abundances of orthologous proteins in the human and rat samples have a certain correlation. The new order of the 981 human orthologs sorted by the abundance of their paired rat proteins in the perfusion-driven urine might approximately represent the abundance order in the pathological condition.
For a given protein, if the abundance ranking increased significantly from the normal urine to the perfusion-driven urine, expression of that protein might increase under pathological conditions compared to other proteins. The ranks of the corresponding rat proteins in the two perfusion-driven urine samples were compared first. The vast majority, 922 proteins (94 %), had ranking changes of less than 300. Therefore, a ranking change of 300 was considered to be significant. In total, 75 of the 981 human orthologs increased in rank by 300 from the normal human urine to the two perfusion-driven urine samples.
The emPAI value is only an approximate estimation of the absolute protein content in a protein mixture [26]. The degree of correlation between orthologous protein abundance was not investigated. Here, we only observed that, for the 75 human orthologs, the rank of their corresponding rat proteins increased significantly in the perfusion-driven urine compared to their rank in the normal urine. We expect the large difference in the abundance ranking of these proteins will indicate their potential to be kidney disease biomarkers.
17.7 Comparison of the Perfusion-Driven Urine Proteomes During Perfusion with and Without Oxygen Supplementation Using LC-MS/MS
The urine proteomes from four independent isolated perfused rat kidneys during perfusion with and without oxygen supplementation were profiled using LC-MS/MS. The samples from two of the rats were profiled with the LTQ Orbitrap Velos platform, which identified 236 and 280 proteins during perfusion with oxygen supplementation and 275 and 281 proteins during perfusion without oxygen supplementation. The samples from the other two rats were profiled with the TripleTOF 5600 platform, which identified 474 and 466 proteins during perfusion with oxygen supplementation and 511 and 527 proteins during perfusion without oxygen supplementation.
The expression of the proteins present during perfusion with oxygen-supplemented medium was compared with expression during perfusion without oxygen supplementation using the label-free quantitative method provided by the SCAFFOLD program. The expression of 39 proteins was significantly increased in all four perfusion-driven urine samples when the kidneys were perfused without oxygen supplementation (p < 0.05, T-test).
These 39 proteins were matched to human orthologs using the same method described above. In total, 33 human orthologs were identified. Because their corresponding rat proteins were increased in the perfusion-driven urine when the kidneys were perfused without oxygen supplementation, these 33 human orthologs were also considered to be the potential human kidney origin proteins in urine.
17.8 Comparison of the Human Kidney Origin Proteins in Urine with Previous Biomarker Studies
A total of 990 non-redundant human orthologs were generated by pooling the perfusion-driven urine proteins that are expressed in the kidney and increased in perfusion-driven urine from oxygen-deficient kidneys. These proteins are potential human kidney origin proteins in urine. Of the 990 kidney origin proteins, there are a total of 428 proteins that may be high-quality potential candidate biomarkers, including kidney origin proteins present in the perfusion-driven urine but not in normal urine, kidney origin proteins present in the perfusion-driven urine but not in the large-scale plasma database, kidney origin proteins that are increased in perfusion-driven urine from oxygen-deficient kidneys, and kidney origin proteins that have a large increase in rank in the perfusion-driven urine compared to normal human urine.
The urinary Protein Biomarker Database was established by comprehensively compiling and manually curating the published literature [27]. A total of 343 candidate biomarkers for human kidney diseases have been collected from the Urinary Biomarker Database [27]. Compared with this database, 67 of the 990 kidney origin proteins have been studied as candidate biomarkers of kidney diseases (Fig. 17.2). Of the 428 high-quality kidney origin proteins, seven proteins have been studied as the candidate biomarkers of kidney diseases.
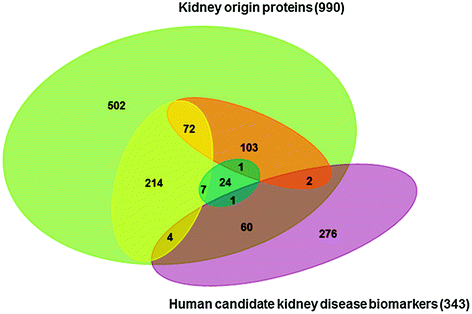

Fig. 17.2
A comparison of the identified kidney origin proteins with previously identified human candidate biomarkers of kidney disease. The yellow oval represents proteins present in perfusion-driven urine but not in normal human plasma. The orange oval represents proteins detected in perfusion-driven urine but not in normal human urine (including human urinary exosomes) or present in human urine but significantly increased in the perfusion-driven urine. The blue oval represents proteins with an increased level in perfusion-driven urine without oxygen supplementation compared to perfusion with oxygen-supplemented medium [28]
However, 923 (93 %) kidney origin proteins have not been studied as candidate biomarkers. Furthermore, few of the 67 kidney origin proteins that were identified as candidates in large-scale differential proteomics experiments have been examined in more detail according to the urinary biomarkers database. One reason why studies examining urinary biomarkers in kidney disease have not been conclusive might be because kidney origin proteins were not examined in detail. These 428 high-quality kidney origin proteins are potential urinary kidney disease markers that should be examined in detail. Because there are hundreds of potentially useful urinary kidney disease markers, combinations of these proteins are likely to be able to differentiate many different kidney conditions.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree







