URL
Content
HKUPP database
Proteome of normal kidney and urine
Urinary exosomes protein database
304 proteins identified from exosomes in normal human urine [11]
MAPU urine dataset
1,543 normal human urinary proteins identified by Adachi et al. [1]
Clinical urine proteomic database
Urinary proteins that were annotated to be associated with diseases by machine learning and text mining methods [6]
Urinary peptide biomarker database
CE-MS results of naturally occurring human urinary peptides in different pathophysiological conditions [14]
Urinary protein biomarker database
A literature-curated database of protein biomarker or biomarker candidates in human and animal urine [13]
19.2 Establishment of the Urinary Protein Biomarker Database
In 2011, we published the Urinary Protein Biomarker Database (UPB database), a manually curated database compiling results of urinary protein biomarker studies from published literature of proteomic studies as well as small-scale experiments such as ELISA and Western blot. Manually, curation ensures the minimum mistakes appear in the process of database establishment.
All of the protein/peptides that were reported to have abundance change under disease conditions are considered as biomarker candidates and collected in this database. No extra filtration of biomarker confidence is used since the aim of building this database was to preserve the original result of literature and make the database as comprehensive as possible. Users can use information such as detection method and sample size displayed in the database to help assess data confidence themselves. Table 19.2 shows data statistics of this database.
Table 19.2
Data statistics of the UPB database after an update in July 2013
Human dataset | Animal dataset | ||||
|---|---|---|---|---|---|
Rat | Mouse | Others | Total | ||
Articles | 348 | 49 | 19 | 8 | 76 |
Records | 993 | 299 | 62 | 18 | 379 |
Diseases | 119 | 29 | 13 | 7 | 49 |
Biomarkers | 819 | 253 | 62 | 18 | 333 |
Proteins | 458 | 161 | 62 | 16 | 239 |
Particularly, a very small proportion of records in this database are ‘negative records,’ in which changes of protein abundance were reported as not statistically significant under disease conditions. These records are included because the same proteins were identified as biomarker candidates for the same diseases by other studies. Including negative data in the biomarker database is important, since it may help researchers to assess data confidence better by analyzing conflict results for the same biomarker candidate.
For each biomarker candidate in this database, the following information was collected (if available): definition and sample size of the disease and control groups, experimental procedures and instrument types, protein information such as fold change, fragment, variant and post-translational modification (PTM), experimental molecular weight and pI. In particular, proteins were queried in the plasma proteome list [5] to infer its origin (Fig. 19.1).
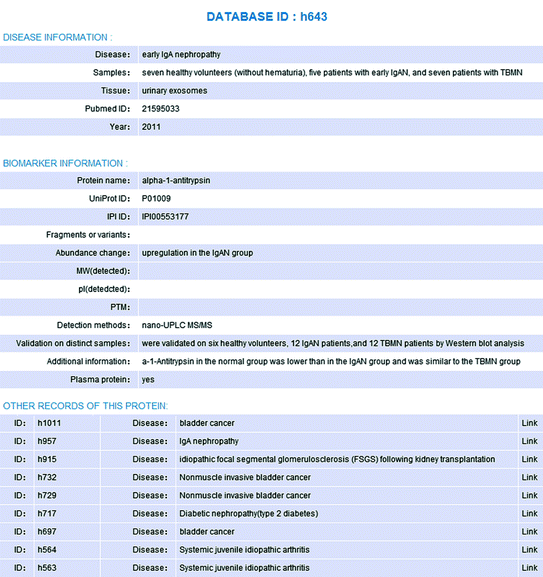

Fig. 19.1




A Webpage displaying records in the UPB database
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree







