Genetics play an important role in the evaluation of the infertile male. The current limitations of classifying the genetic contribution to male infertility and the importance of phenotyping men are discussed, and the core concepts necessary to interpret most genetic studies are reviewed. The current genetic assays used clinically are discussed in detail. The use and interpretation of the cystic fibrosis transmembrane receptor assay are examined in the context of men with clinical bilateral absence of the vas deferens, a karyotype and Klinefelter syndrome, and Y chromosome microdeletions. The role of hormones and epigenetics in evaluating the genetic reproductive potential of men is discussed briefly. A summary of what the field might look like in 2034 is presented.
Key points
- •
Much of idiopathic male infertility is likely to have a genetic cause.
- •
Men who have nonobstructive azoospermia or severe oligospermia with total motile count less than 5 million should have a karyotype and Y chromosome microdeletion.
- •
Klinefelter syndrome (47,XXY) is the most common chromosomal abnormality with a frequency of 1:600 males and has a wide spectrum of clinical presentation.
- •
Men with an AZFa , AZFb , AZFb/c microdeletion uniformly have complete absence of spermatogenesis.
- •
If a male has congenital bilateral absence of the vas deferens, it is critical to offer him and his partner genetic testing for cystic fibrosis mutations as well as genetic counseling.
Introduction
Approximately 1 in 6 couples in the Western world is not able to conceive spontaneously after 1 year of unprotected intercourse; in nearly half of these couples, the male partner has 1 or more semen parameters below the WHO cutoffs for normozoospermia. Although the sequencing of the human genome in 2003 heralded a new era of genetic medicine, it will likely take decades to realize the potential of this project. Male infertility, in part due to the nature of the condition, remains largely unexplained. The cause of most cases of male infertility or subfertility remains unknown; monogenic disorders (eg, cystic fibrosis [CF], Kallman syndrome), cytogenetic abnormalities (eg, Klinefelter syndrome [KS; 47,XXY]), and Y chromosome deletions account for only up to 30% of cases. The proportion of the remaining male factor cases that can be attributed to genetic causes is currently unknown, but it is likely that aberrations in many additional genes underlie a significant proportion of male infertility/subfertility because sperm production requires the coordinated action of thousands of genes, and knocking out any 1 of hundreds of genes in mice results in subfertility phenotypes in males. However, discovering such genes in humans has proved challenging.
Based on studies of animal models, however, it is likely that genetic variation that alters gene expression or function accounts for a significant proportion of male subfertility. For example, knock outs of or mutations in hundreds of genes cause subfertility phenotypes in male mice. This is not surprising given that sperm development and maturation require the coordinated action of thousands of genes. However, identifying the variation and specific genes that are essential for reproductive success in humans has been extremely challenging for 2 reasons. First, because of the nature of the condition, it is virtually impossible to conduct genome-wide family-based studies of infertility, approaches that have been successful for identifying genes for many conditions with monogenic, and even some with complex genetic causes. Second, male infertility is a heterogeneous condition that can result from aberrations of many different genes. This is due in part to strong selection pressure against transmission of these genetic variants. As a result, candidate gene association studies (or even genome-wide association studies [GWAS]) of cases (infertile) and control (fertile) men would not likely be successful because only a small proportion of the cases are expected to share the same genetic abnormality. This is shown by the relative paucity of specific genetic variants and genes that are robustly associated with male infertility.
Our lack of success in explaining approximately 50% to 70% of male infertility is nowhere more apparent than in our interactions with infertile men. These men want an answer to what caused their infertility. Currently, we cannot provide this in most instances. Furthermore, technological advances such as intracytoplasmic sperm injection (ICSI) and microsurgical testicular sperm extraction (microTESE) allow us to bypass the problem and bring with them another set of questions from patients that we cannot answer. When considering ICSI, many patients want to know what are the chances they will pass on the genetic cause of their infertility to their offspring, as well as the potential for nonreproductive effects from these genes. These are questions that currently cannot completely answered completely. Although studies suggest that assisted reproductive technologies do not seem to result in a significantly higher rate of birth defects after risk factors such as maternal age are controlled for, the role of sperm quality in reproduction is just beginning to be unraveled.
In 2012, Kong and colleagues published a seminal paper in Nature showing that the de novo mutation rate for each generation is driven largely by paternal age with paternal sperm mutation rate doubling for every 16-year increase in paternal age. Increased paternal mutations from advancing age of fathers explained 30% of the increase in autism and schizophrenia over the time period of this study. The mechanism driving this is believed to be increased de novo mutations resulting from decreased fidelity of DNA replication in spermatogenesis with advancing paternal age. These mutations result in a higher mutation rate in sperm, which are then passed on to offspring and can manifest as diseases such as schizophrenia or autism.
Studies such as that of Kong and colleagues and recent work by Wang and colleagues, which sequenced the entire genome of individual sperm, herald a paradigm shift in our ability to develop the next generation of genetic tools to understand and possibly treat the underlying cause of male infertility. Tools such as this provide the ability to interrogate the reproductive potential of individual sperm, unfortunately, at this time, this cannot be done without destroying them. However, this technology holds incredible potential to determine the reproductive potential of an individual sperm.
Voltaire said, “with great power comes great responsibility.” In many ways, ICSI and microTESE have given us incredible power to treat male infertility. With this power, comes the ethical and moral responsibility to understand the genetic causes of male infertility for our patients and their offspring. Much of the potential of the Human Genome Project will be brought to bear on the genetic causes of male infertility.
This article examines some basic concepts that are prerequisite to any examination of the genetic causes of male infertility and reviews who should be evaluated and the current tools for genetic evaluation as well as their limitations. An overview of state of the art research in the field and what the landscape will look like in 2034 are presented.
Introduction
Approximately 1 in 6 couples in the Western world is not able to conceive spontaneously after 1 year of unprotected intercourse; in nearly half of these couples, the male partner has 1 or more semen parameters below the WHO cutoffs for normozoospermia. Although the sequencing of the human genome in 2003 heralded a new era of genetic medicine, it will likely take decades to realize the potential of this project. Male infertility, in part due to the nature of the condition, remains largely unexplained. The cause of most cases of male infertility or subfertility remains unknown; monogenic disorders (eg, cystic fibrosis [CF], Kallman syndrome), cytogenetic abnormalities (eg, Klinefelter syndrome [KS; 47,XXY]), and Y chromosome deletions account for only up to 30% of cases. The proportion of the remaining male factor cases that can be attributed to genetic causes is currently unknown, but it is likely that aberrations in many additional genes underlie a significant proportion of male infertility/subfertility because sperm production requires the coordinated action of thousands of genes, and knocking out any 1 of hundreds of genes in mice results in subfertility phenotypes in males. However, discovering such genes in humans has proved challenging.
Based on studies of animal models, however, it is likely that genetic variation that alters gene expression or function accounts for a significant proportion of male subfertility. For example, knock outs of or mutations in hundreds of genes cause subfertility phenotypes in male mice. This is not surprising given that sperm development and maturation require the coordinated action of thousands of genes. However, identifying the variation and specific genes that are essential for reproductive success in humans has been extremely challenging for 2 reasons. First, because of the nature of the condition, it is virtually impossible to conduct genome-wide family-based studies of infertility, approaches that have been successful for identifying genes for many conditions with monogenic, and even some with complex genetic causes. Second, male infertility is a heterogeneous condition that can result from aberrations of many different genes. This is due in part to strong selection pressure against transmission of these genetic variants. As a result, candidate gene association studies (or even genome-wide association studies [GWAS]) of cases (infertile) and control (fertile) men would not likely be successful because only a small proportion of the cases are expected to share the same genetic abnormality. This is shown by the relative paucity of specific genetic variants and genes that are robustly associated with male infertility.
Our lack of success in explaining approximately 50% to 70% of male infertility is nowhere more apparent than in our interactions with infertile men. These men want an answer to what caused their infertility. Currently, we cannot provide this in most instances. Furthermore, technological advances such as intracytoplasmic sperm injection (ICSI) and microsurgical testicular sperm extraction (microTESE) allow us to bypass the problem and bring with them another set of questions from patients that we cannot answer. When considering ICSI, many patients want to know what are the chances they will pass on the genetic cause of their infertility to their offspring, as well as the potential for nonreproductive effects from these genes. These are questions that currently cannot completely answered completely. Although studies suggest that assisted reproductive technologies do not seem to result in a significantly higher rate of birth defects after risk factors such as maternal age are controlled for, the role of sperm quality in reproduction is just beginning to be unraveled.
In 2012, Kong and colleagues published a seminal paper in Nature showing that the de novo mutation rate for each generation is driven largely by paternal age with paternal sperm mutation rate doubling for every 16-year increase in paternal age. Increased paternal mutations from advancing age of fathers explained 30% of the increase in autism and schizophrenia over the time period of this study. The mechanism driving this is believed to be increased de novo mutations resulting from decreased fidelity of DNA replication in spermatogenesis with advancing paternal age. These mutations result in a higher mutation rate in sperm, which are then passed on to offspring and can manifest as diseases such as schizophrenia or autism.
Studies such as that of Kong and colleagues and recent work by Wang and colleagues, which sequenced the entire genome of individual sperm, herald a paradigm shift in our ability to develop the next generation of genetic tools to understand and possibly treat the underlying cause of male infertility. Tools such as this provide the ability to interrogate the reproductive potential of individual sperm, unfortunately, at this time, this cannot be done without destroying them. However, this technology holds incredible potential to determine the reproductive potential of an individual sperm.
Voltaire said, “with great power comes great responsibility.” In many ways, ICSI and microTESE have given us incredible power to treat male infertility. With this power, comes the ethical and moral responsibility to understand the genetic causes of male infertility for our patients and their offspring. Much of the potential of the Human Genome Project will be brought to bear on the genetic causes of male infertility.
This article examines some basic concepts that are prerequisite to any examination of the genetic causes of male infertility and reviews who should be evaluated and the current tools for genetic evaluation as well as their limitations. An overview of state of the art research in the field and what the landscape will look like in 2034 are presented.
Phenotype definitions
Studying the genetics of male infertility is complex because many of the tools of genetic analysis such as linkage mapping, family studies, and complex pedigree analysis are rendered useless by the nature of the condition. Furthermore, male infertility exists on a spectrum and is likely the result of the contribution of 100s if not 1000s of genes to a man’s overall reproductive potential. To study this or any other genetic condition, accurate phenotyping is essential. To determine the precise genetic cause of male fertility, robust definitions that can clearly differentiate men into similar groups for analysis are essential. If this often overlooked but critical step cannot be completed, our efforts are doomed to failure. Although significant progress is being made in genomic, proteomic, and metabolomics biomarkers of male infertility, the limiting factor in this work is lack of accurate phenotyping of these men from a clinical and molecular standpoint ( Table 1 ). Another key component of accurately phenotyping men is to define accurate inclusion and exclusion criteria to establish a uniform cohort of men for analysis ( Table 2 ).
| Demographic data | Semen analysis |
| Age (y) | Volume (mL) |
| Partner’s age (y) | Sperm count |
| Race/ethnicity | % Motility |
| Body mass index (kg/m 2 ) | Total motile count |
| Hormones | % Progressive motility |
| Follicular stimulating hormone (mIU/mL) | Average velocity |
| Luteinizing hormone (mIU/mL) | Mean amplitude of lateral head movement |
| Total and free testosterone (ng/dL) | Linearity |
| Clinical | Beat frequency |
| Months of infertility | Morphology (% normal) |
| Female factor present in partner | % Head defects |
| Anatomic | % Neck/midpiece defects |
| Testis longitudinal axis (cm) | % Cyoplasmic defects |
| Nonsevere varicocele (grade I or II) | % Tail defects |
| A. Inclusion Criteria | |
| Men aged 18–65 y in a committed relationship | |
| No previous paternity | |
| B. Exclusion Criteria | |
| Medical history | Cryptorchidism/orchidopexy Severe testicular trauma or torsion Previous inguinal surgery Vasectomy Radical pelvic surgery Chemotherapy Pelvic external beam radiotherapy/brachytherapy Cancer (other than nonmelanoma skin cancer) HIV/AIDS Mumps orchitis CF or CBAVD Spinal cord injury |
| Hormonal | Hypogonadotropic hypogonadism Hyperprolactinemia Hyper or hypothyroidism Diabetes mellitus with HbA1C >10% Exogenous steroid use |
| Anatomic | Grade III varicocele Severe phimosis Presence of testicular mass Buried penis due to morbid obesity |
| Genetic | AZF microdeletion Klinefelter syndrome Intersex disorder CFTR mutation |
| Semen analysis | Seminal hypovolemia (volume<1.5 mL) |
| Sexual history | Ejaculatory dysfunction |
Previous investigators have focused on men with nonobstructive azoospermia (NOA) to identify a pure phenotype with a uniform condition. Although this approach is appealing in that NOA is certainly a reproducible end point and clearly defines a population of patients, it has not been successful in identifying genetic causal variants that explain large portions of male infertility. Much of this is believed to be due to racial and ethnic differences in genetic carrier frequencies and the 100s of genetic defects that can result in an NOA phenotype. Given that most men do not realize their full reproductive potential, that birth outcomes are also dependent on female factors, and that semen analyses are notoriously variable, NOA provides an attractive phenotypic definition for male infertility. The problem with using men with NOA as a phenotypic definition of male factor infertility is that significant numbers of men with causal genetic variants that contribute to subfertility or severe oligozoospermia through genetic pathways distinct from those that cause NOA may be missed. Furthermore, because NOA is only a small subgroup of men with male infertility, it is unclear if understanding the genetic causes of NOA will translate directly into deciphering other aspects of male infertility.
Alternative phenotypic definitions for male infertility have their own problems and limitations as well. Specifically, using patient self-reports of their fertility is problematic and, if used, needs to be done in a validated and controlled manner; it will only work in specific populations where men realize their true genetic reproductive potential. Many couples now seek assisted reproductive technologies before attempting to conceive for 12 months. Alternatively, investigators have relied on semen analyses to define groups of men with oligozoospermia, but variability in semen analyses mandates use of multiple semen samples to define these groups of men. Case-control definitions are also problematic because semen analyses parameters, such as total motile counts, are quantitative continuously distributed traits that show large intraindividual and interindividual variation. Thus, dichotomizing total motile count would fail to detect an overall reduction in sperm count caused by a genetic factor, unless the cutoff point for the case definition was set very low. Finding accurate controls for these studies has also proved to be problematic.
One alternative to just relying on NOA or oligozoospermia on a semen analysis is to define more robust clinical phenotypes that tie in other relevant pieces of clinical information such as the physical examination and hormone levels ( Table 3 ). Male and female reproductive hormone levels are an integral part of an infertility evaluation and frequently change clinical management. For men, the level of follicular stimulating hormone (FSH) is often more predictive of spermatogenesis capacity than a semen analysis, assuming absence of azoospermia, and luteinizing hormone (LH) and testosterone (T) often identify treatable hypoandrogenism. Multiple studies have demonstrated that levels of FSH, LH, T, and anti-Mullerian hormone (AMH) in men have significant heritability (56%–90%). Previous studies of twins have suggested that levels of these hormones are heritable, but further genetic studies on these important biomarkers of reproductive health are lacking.
| Male Infertility Metric | Rationale for Measurement |
|---|---|
| Demographic | |
| Age (y) | Semen quality decreases with age >25 y |
| Race/ethnicity | Significant racial variability in male infertility prevalence, care seeking behavior, and semen analysis profiles mandates adjustment by race |
| Body mass index (BMI) (kg/m 2 ) | Increasing BMI is associated with declining semen quality |
| Partner age (y) | Increasing female age is associated with decreased fertility |
| Hormones | |
| Follicular stimulating hormone (FSH) (mIU/mL) | FSH correlates directly with spermatogenesis potential and is significantly less variable and more heritable than the parameters of a semen analysis |
| Luteinizing hormone (mIU/mL) | Indicates adequacy of Leydig cell function to maintain adequate testosterone levels for spermatogenesis |
| Free testosterone (ng/dL) | Adequate free testosterone is necessary to optimize human spermatogenesis and does not always correlate with total testosterone |
| Total testosterone (ng/dL) | Total testosterone <280 ng/dL has sensitivity of 91.0% and specificity of 73.7% for low free testosterone |
| Semen Analysis | |
| Volume (mL) | Seminal hypovolemia (<1.5 mL) indicates obstructive azoospermia or retrograde ejaculation, not spermatogenic failure |
| Sperm count (millions of sperm) | Pregnancy rates decline with decreasing sperm counts |
| Total motile count (TMC) (millions of sperm) | Clinically, TMC is used to determine the severity of male factor infertility and to guide clinical treatment. Pregnancy rates are believed to decline linearly with reduced TMC <15 million |
| % Motility | Decreases in motility can indicate genetic defects in spermatogenesis that can result in populations of immotile sperm |
| % Normal morphology by Kruger strict criteria | Previous work has found that genes associated with reduced reproductive potential are also associated with specific morphologic defects. Decreased % of normal forms may be associated with decreased fertility and is directly related to the quality of the germinal epithelium |
| % Head defects | Previous work has found that genes associated with reduced reproductive fitness are also associated with sperm head defects |
| Anatomic | |
| Testis longitudinal axis | 80% of testicular volume is composed of the seminiferous tubules, where spermatogenesis occurs. Thus, testicular size is directly proportional to reproductive fitness and does not have the variability seen in semen analyses |
| Presence/side/grade of varicocele | Varicoceles are associated with oligoasthenoteratospermia and are found in up to 50% of men presenting to infertility clinics. However, they are often an incidental finding and are typically not causative of severe defects in spermatogenesis. Thus, men with severe (grade III) varicoceles are excluded because the genetic factors causing varicoceles are unknown but are believed to differ from those causing defects in spermatogenesis |
| Clinical * | |
| Months of infertility at time of evaluation | Reproduction is an inefficient process, even in fertile couples, with chances of fertilization approaching 20% under ideal conditions. Severity of male factor infertility correlates linearly with length of time to natural conception without assisted reproductive technologies. Infertility is defined clinically as the lack of pregnancy after 1 y of attempts at pregnancy |
| Months until natural conception | See above |
| Months until conception with intrauterine insemination | Chances of pregnancy with intrauterine insemination are roughly 15%–20% per cycle. Because intrauterine insemination success usually requires a TMC >5 million and is proportional to sperm function, this can be used as a surrogate for reproductive fitness |
Genetics background
Basic Genetics
The central dogma of biology dictates that DNA makes RNA, which makes protein. Proteins form the building blocks of life. The basic building blocks of genes, the genetic code, consists of 4 deoxyribonucleotides (adenylic acid [A], guanylic acid [G], thymidylic acid [T], and cytidylic acid [C]). Two strands of DNA are joined together to form a double helix with A binding to T and G binding to C. DNA consists of introns, sections of DNA that do not code for proteins and exons, and sections of DNA that code for proteins. Although only a small fraction of DNA codes for proteins, recent insights from the ENCODE study have revealed that the intervening DNA is not random noise but serves to regulate the exons or coding segments. The DNA of each gene is transcribed to make mRNA. During translation, each 3-unit nucleotide codon is translated by the ribosome to make a specific amino acid. Sequences of amino acids then make specific proteins, the functional end product of each gene.
DNA is tightly packaged in the nucleus of cells. It is set in a background of histone proteins, stacked and compacted to form each of the 46 chromatids, which consist of a short (p) arm and a long (q) arm. One of the chromatids is of paternal origin and 1 is of maternal origin. These chromatids make up the diploid genome which consists of 22 pairs or autosomes numbered from largest to smallest and 1 pair of sex chromosomes (X/Y or XX) ( Fig. 1 ).
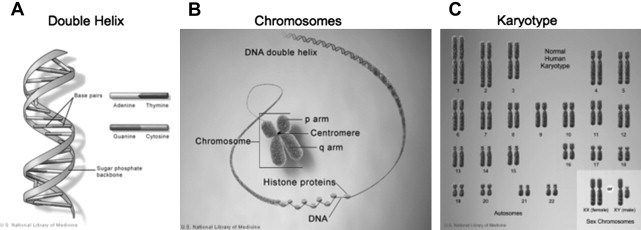
DNA is replicated in the process of mitosis and meiosis. Mitosis occurs in all cells and precisely replicates the DNA to produce 2 genetically identical diploid daughter cells from each mother cell. Meiosis occurs only in germ cells and involves a process of recombination and reduction in chromosome number to a haploid spermatozoa or oocyte. Fusion of an oocyte and a spermatozoa result in restoration of the diploid number of chromatids.
DNA mutations
A mutation is an alteration in DNA that can be passed from parent to daughter cells. There is a critical distinction between somatic mutations and germline mutations. Somatic mutations are passed from mother to daughter cells, but not passed on to the next generation. The rate of de novo germline mutations is not insignificant and tends to increase as people age. Both germline and somatic mutations may result in a change in the amino acid sequence of a protein or the length of genes (insertions or deletions). In this article, the discussion of genetic inheritance focuses on germline genetic disorders and this model of inheritance.
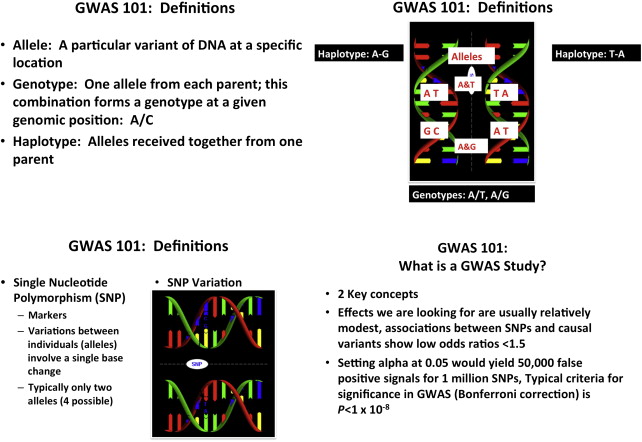
DNA polymorphisms
Polymorphisms are alterations in the DNA found in at least 1% of the population. Generally speaking, DNA polymorphisms do not cause disease but may alter the risk or severity of disease. There are several types of polymorphisms. The most common and most relevant for male infertility are single nucleotide polymorphisms (SNPs), which occur in up to 1 in 100 base pairs for some genes and typically do not cause disease. SNPs are specific areas of DNA that vary between individuals in a population. An allele is a specific variant of DNA at a specific location, whereas a genotype is the alleles an individual received from each parent at a given genomic position such as A/C. A haplotype is the alleles that were each received together from 1 parent. GWAS attempt to determine whether the genotypes of certain SNPs are associated with complex diseases. These studies generate massive amounts of data and are complex to interpret but are statistically relatively straightforward although computationally intensive; they rely on millions of t tests to examine the association of the genetic predictor (SNP genotypes) with the outcome of interest. Examination of millions of SNPs in a given study often results in stringent criteria for genome-wide significance ( P <1 × 10 −8 ) after correcting for multiple comparisons ( Fig. 2 ). GWAS, when properly performed, adequately powered and correctly interpreted, may have the power to yield insight into complex diseases such as male infertility.
Genetic Disorders
Genetic disorders can be divided into single gene disorders or mendelian disorders, chromosomal disorders, and nonmendelian genetic disorders. All the examples mentioned briefly here are discussed in detail in the following sections. Single gene or mendelian disorders are caused by a mutant allele or pair of alleles at a single genetic locus. These mutant alleles may either be inherited from parents or occur de novo in spermatozoa or oocytes. Regardless of where they come from, once present, all these mendelian disorders are passed on to offspring in 1 of several standard modes of inheritance. Autosomal dominant mutations are expressed with the inheritance of a single mutant allele, whereas autosomal recessive mutations require the disease-causing mutation to be present on both alleles of a gene. CF is a classic example of an autosomal recessive mutation. X-linked disorders cause disease in men (46,XY) with the mutation and in women who inherit 2 copies of the X-linked mutation. Thus, these diseases affect men more than women. Kallman syndrome is an example of an X-linked disorder.
Chromosomal disorders are caused by the loss, gain, or abnormal arrangement of 1 or more of the 46 chromosomes. Although most chromosomal disorders are de novo events that result from significant mutations in the parent germ cells, they often demonstrate a modified pattern of mendelian inheritance. These disorders can be classified as either numerical/structural or microscopic/submicroscopic. There are 2 categories of numerical chromosomal abnormalities: (1) polyploidy, a chromosomal number that is a multiple of 23 in which there are extra copies of all chromosomes; (2) aneuploidy, a gain or loss of 1 or more chromosomes. Aneuploidy is typically denoted as the number of extra or missing copies and the chromosome; for example, trisomy 21. Aneuploidy is significantly more common than polyploidy. Mosaicism results when individuals have tissues consisting of a mixture of cell lineages with different chromosomal complements. A classic example of a numerical aneuploid chromosomal disorder where mosaicism is common is KS with a karyotype of 47,XXY.
Microscopic or submicroscopic chromosomal disorders result from a loss, gain, or rearrangement of material within a chromosome or between chromosome. The key distinction between these and numerical chromosomal abnormalities is that only a piece of the chromosome is affected, not the entire chromosome. A common mechanism for these disorders is reconfiguration of blocks of DNA or low copy repeats, which are 10 to 400 kb long, have nearly identical sequences, and are dispersed throughout the chromosome accounting for 5% of the human genome. A classic example of this is deletions of part of the Y chromosome resulting in microdeletions leading to male infertility, the so-called azoospermia factor (AZF) disorders.
Nonmendelian disorders account for most human disease. Study of these diseases is significantly more complex than for mendelian disorders. GWAS have been the mainstay used to investigate complex, polygenic, nonmendelian diseases (see Fig. 2 ). Other inheritance patterns exist such as expansion of trinucleotide repeats, mitochondrial inheritance, genomic imprinting, and uniparental disomy but these are beyond the scope of this article. Studying spermatogenesis is complex and requires understanding how 1000s of genes operate together and the development of new tools to examine complex nonmendelian traits such as GWAS.
Current genetic tools
Spermatogenesis involves the coordinated action of 1000s of genes. Although any number of these could make excellent targets for diagnostic tests of male factor infertility or subfertility, only a small handful of genetic variants have been clearly linked to spermatogenic failure in a robust and reproducible manner.
Congenital Bilateral Absence of the Vas Deferens
Congenital bilateral absence of the vas deferens (CBAVD) occurs in approximately 1% of infertile men and is diagnosed on physical examination, prompting subsequent genetic testing. Men with this condition present with obstructive azoospermia, absence of the vas deferens, and possibly absence of the distal part of the epididymis, hypoplastic seminal vesicles, and consequent seminal hypovolemia (<1 mL) and an acidic ejaculate (pH 6.5–7.0). CBAVD is found in all patients with clinical CF and CBAVD without other clinical manifestations of CF is believed to result in people with mutations that confer at least some functional forms of the gene that causes CF when it is completely absent.
CF affects 1:1600 people of northern European descent and genetic testing must account for ethnicity to identify the 850 or so genetic variants known to cause CF. Obstructive pulmonary disease caused by thickened epithelial secretions is the defining feature of clinical CF; pancreatic exocrine failure from the same mechanism is also common. Absence of the vas deferens occurs in all males with clinical CF. Clinical CF requires inheritance of maternal and paternal CF genes. The CF gene encodes the cystic fibrosis transmembrane conductance regulator (CFTR), a protein crucial for the maintenance of viscosity through optimal sodium and chloride balance in epithelial secretions. If only 1 copy of an abnormal CFTR gene is present along with another normal copy, the patient is a carrier and pancreatic and respiratory function are unaffected. The severity of the phenotypic picture of CF, from carrier to clinical CF, depends on the functionality of the copies of the CFTR genes that individuals inherit from their parents. The least severe form of CF is CBAVD, where the CFTR protein allows for adequate pancreatic and respiratory function but results in vasal agenesis. Although the vas, epididymis, and seminal vesicles are of mesonephric origin, they become atretic in the later stages of development, indicating that the mesopnephric ducts are embryologically normal and men with CBAVD have normal renal units.
Although more than 1500 mutations can cause CF and CBAVD, a 3 base pair deletion, deltaF508, is the most common mutation found in northern Europeans with CF and CBAVD. deltaF508 is a severe mutation and the homozygous state results in clinical CF. In men with CBAVD, complete genome sequencing results in detection of 90% of abnormal CFTR alleles (the other 10% are presumed but not detectable); 88% carry a severe mutation (absent CFTR function) in combination with an allelic mild mutation that preserves some CFTR function. The most frequent mutation detected is deltaF508 (24%) and the second most common is IVS8-T5 (17%). T5 causes mild CFTR malfunction and is present in up to 5% of the general population. The most frequent genetic combination in patients with CBAVD was deltaF508 in trans to IVS8-T5 (16.5%). Most other CFTR mutations were at a frequency of 3% or less. Unilateral absence of the vas deferens should also be evaluated with renal ultrasonography because many of these men have renal agenesis or ectopia. Another variant of this is congenital nonunion of the vas deferens with the epididymis, which is poorly understood and may lend itself to microsurgical reconstruction in some instances.
If no mutations in CFTR are discovered in the male, another possible cause of CBAVD is from abnormal differentiation of the mesonephric ducts before week 7 resulting in unilateral renal agenesis or ectopy and CBAVD. This scenario occurs in its severe form as Potter syndrome, is unrelated to CF, has an unknown genetic basis, and warrants renal ultrasonography in men with CBAVD to identify this entity.
Perhaps the most critical portion of an evaluation of CBAVD is workup of the female partner for CFTR mutations. According to the American Urological Association (AUA) Best Practice Policy Committee’s Report from 2010 on the Evaluation of the Azoospermic Male , “Testing for cystic fibrosis transmembrane conductance regulator abnormalities should include at minimum a panel of common point mutations and the 5T allele. Gene sequencing may be considered in couples where the wife is a carrier and the husband with congenital bilateral absence of the vasa deferentia tests negative on a routine panel of cystic fibrosis transmembrane conductance regulator mutations.” ( https://www.auanet.org/common/pdf/education/clinical-guidance/Male-Infertility-b.pdf ). Referral to a genetic counselor is a critical component of this process.
Men with CBAVD and CF should undergo genetic screening and are then candidates for microsurgical or percutaneous sperm aspiration procedures or testicular sperm extraction for use in conjunction with ICSI. Preimplantation genetic screening should be done if the patient’s partner harbors a CFTR mutation, resulting in a 25% chance of offspring inheriting abnormal alleles from both parents and developing clinical CF. The key points of the evaluation of a man presenting with CBAVD are summarized in Table 4 .





